Gt(ROSA)26Sortm3(rtTA,tetO-cre/ERT)Nat
Targeted Allele Detail
|
|
| Symbol: |
Gt(ROSA)26Sortm3(rtTA,tetO-cre/ERT)Nat |
| Name: |
gene trap ROSA 26, Philippe Soriano; targeted mutation 3, Jeremy Nathans |
| MGI ID: |
MGI:3849063 |
| Synonyms: |
R26rtTACreER |
| Gene: |
Gt(ROSA)26Sor Location: Chr6:113044389-113054205 bp, - strand Genetic Position: Chr6, 52.73 cM
|
| Alliance: |
Gt(ROSA)26Sortm3(rtTA,tetO-cre/ERT)Nat page
|
|
| Germline Transmission: |
Earliest citation of germline transmission:
J:155417
|
| Parent Cell Line: |
Not Specified (ES Cell)
|
| Strain of Origin: |
129
|
|
| Allele Type: |
|
Targeted (Inducible, Recombinase, Transactivator) |
| Inducer: |
|
tamoxifen and tetracycline |
| Mutation: |
|
Insertion
|
| |
|
Gt(ROSA)26Sortm3(rtTA,tetO-cre/ERT)Nat expression driven by
1 gene
Knock-in expression driven by:
| Organism |
Driver Gene |
Note |
| bacteria |
tetO |
|
|
| |
|
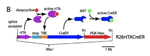
Mutation details: The construct consists of the rtTA coding region preceded by a splice acceptor so that it can be spliced onto the 5' exon (exon1) of the endogenous ROSA26 transcript, and is followed by six repeats of an SV40 transcriptional terminator signal ("stop"). In the presence of doxycycline, the rtTA protein activates transcription from the seven head-to-tail Tet response elements immediately 5' of a minimal cytomegalovirus immediate early promoter (TRE) of the CreER fusion protein open reading frame (followed by the bovine growth hormone 3' UTR and transcriptional terminator region), leading to expression of CreER.
(J:155417)
|
|
Generation of the Gt(ROSA)26Sortm3(rtTA,tetO-cre/ERT)Nat allele |
|
|
|
Activity: |
 Tissue activity of this recombinase allele
|
|
Driver:
|
tetO
(bacteria)
|
| |
|
|
View phenotypes and curated references for all genotypes (concatenated display).
|
|
|
| Original: |
J:155417 Badea TC, et al., New mouse lines for the analysis of neuronal morphology using CreER(T)/loxP-directed sparse labeling. PLoS One. 2009;4(11):e7859 |
| All: |
4 reference(s) |
|
 Analysis Tools
Analysis Tools





