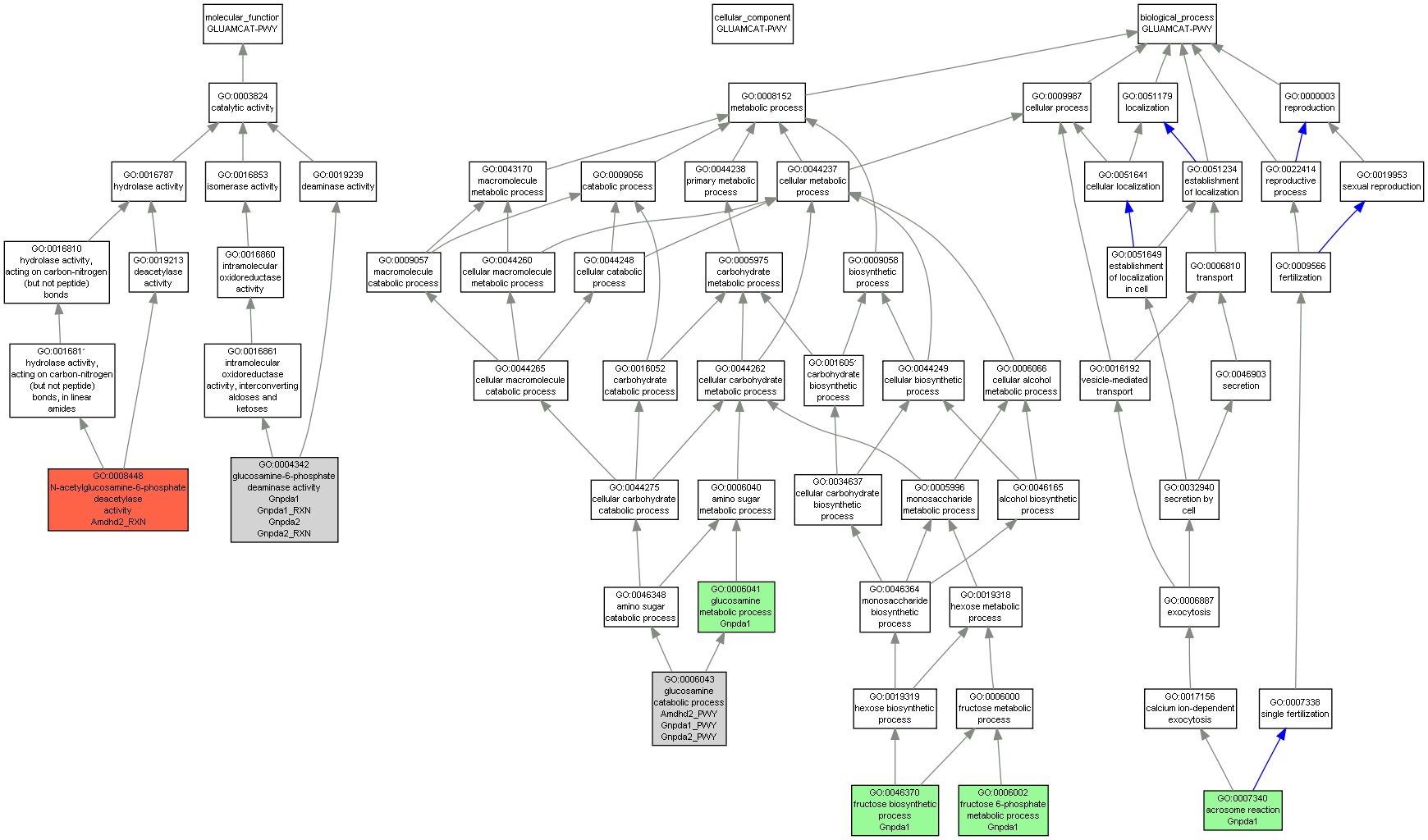
| Category | ID | Term | Mouse gene |
Evidence | Color Key |
| Molecular Function | GO:0004342 | glucosamine-6-phosphate deaminase activity |
Gnpda1 |
MGI_noIEA | color key |
| Molecular Function | GO:0004342 | glucosamine-6-phosphate deaminase activity |
Gnpda2 |
MGI_noIEA | color key |
| Molecular Function | GO:0004342 | glucosamine-6-phosphate deaminase activity |
Gnpda1_RXN |
EC-NUMBER | color key |
| Molecular Function | GO:0004342 | glucosamine-6-phosphate deaminase activity |
Gnpda2_RXN |
EC-NUMBER | color key |
| Molecular Function | GO:0008448 | N-acetylglucosamine-6-phosphate deacetylase activity |
Amdhd2_RXN |
EC-NUMBER | color key |
| Biological Process | GO:0007340 | acrosome reaction |
Gnpda1 |
MGI_noIEA | color key |
| Biological Process | GO:0006002 | fructose 6-phosphate metabolic process |
Gnpda1 |
MGI_noIEA | color key |
| Biological Process | GO:0046370 | fructose biosynthetic process |
Gnpda1 |
MGI_noIEA | color key |
| Biological Process | GO:0006043 | glucosamine catabolic process |
Amdhd2_PWY |
EC-NUMBER | color key |
| Biological Process | GO:0006043 | glucosamine catabolic process |
Gnpda1_PWY |
EC-NUMBER | color key |
| Biological Process | GO:0006043 | glucosamine catabolic process |
Gnpda2_PWY |
EC-NUMBER | color key |
| Biological Process | GO:0006041 | glucosamine metabolic process |
Gnpda1 |
MGI_noIEA | color key |