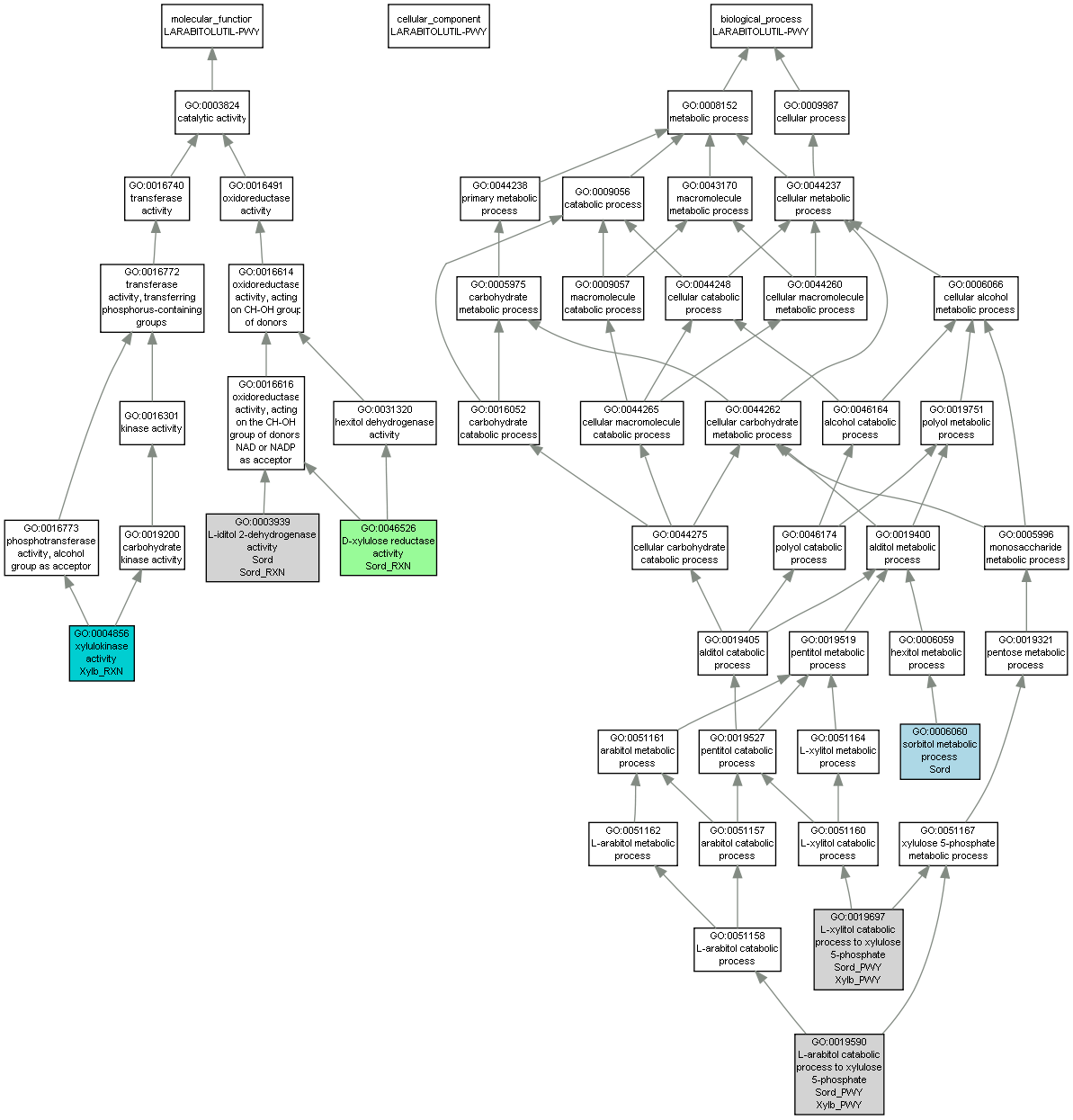
| Category | ID | Term | Mouse gene |
Evidence | Color Key |
| Molecular Function | GO:0046526 | D-xylulose reductase activity |
Sord_RXN |
AUTOMATED-NAME-MATCH | color key |
| Molecular Function | GO:0003939 | L-iditol 2-dehydrogenase activity |
Sord |
MGI_noIEA | color key |
| Molecular Function | GO:0003939 | L-iditol 2-dehydrogenase activity |
Sord_RXN |
EC-NUMBER | color key |
| Molecular Function | GO:0004856 | xylulokinase activity |
Xylb_RXN |
EC-NUMBER | color key |
| Biological Process | GO:0019590 | L-arabitol catabolic process to xylulose 5-phosphate |
Sord_PWY |
AUTOMATED-NAME-MATCH | color key |
| Biological Process | GO:0019590 | L-arabitol catabolic process to xylulose 5-phosphate |
Sord_PWY |
EC-NUMBER | color key |
| Biological Process | GO:0019590 | L-arabitol catabolic process to xylulose 5-phosphate |
Xylb_PWY |
EC-NUMBER | color key |
| Biological Process | GO:0019697 | L-xylitol catabolic process to xylulose 5-phosphate |
Sord_PWY |
AUTOMATED-NAME-MATCH | color key |
| Biological Process | GO:0019697 | L-xylitol catabolic process to xylulose 5-phosphate |
Sord_PWY |
EC-NUMBER | color key |
| Biological Process | GO:0019697 | L-xylitol catabolic process to xylulose 5-phosphate |
Xylb_PWY |
EC-NUMBER | color key |
| Biological Process | GO:0006060 | sorbitol metabolic process |
Sord |
MGI_noIEA | color key |