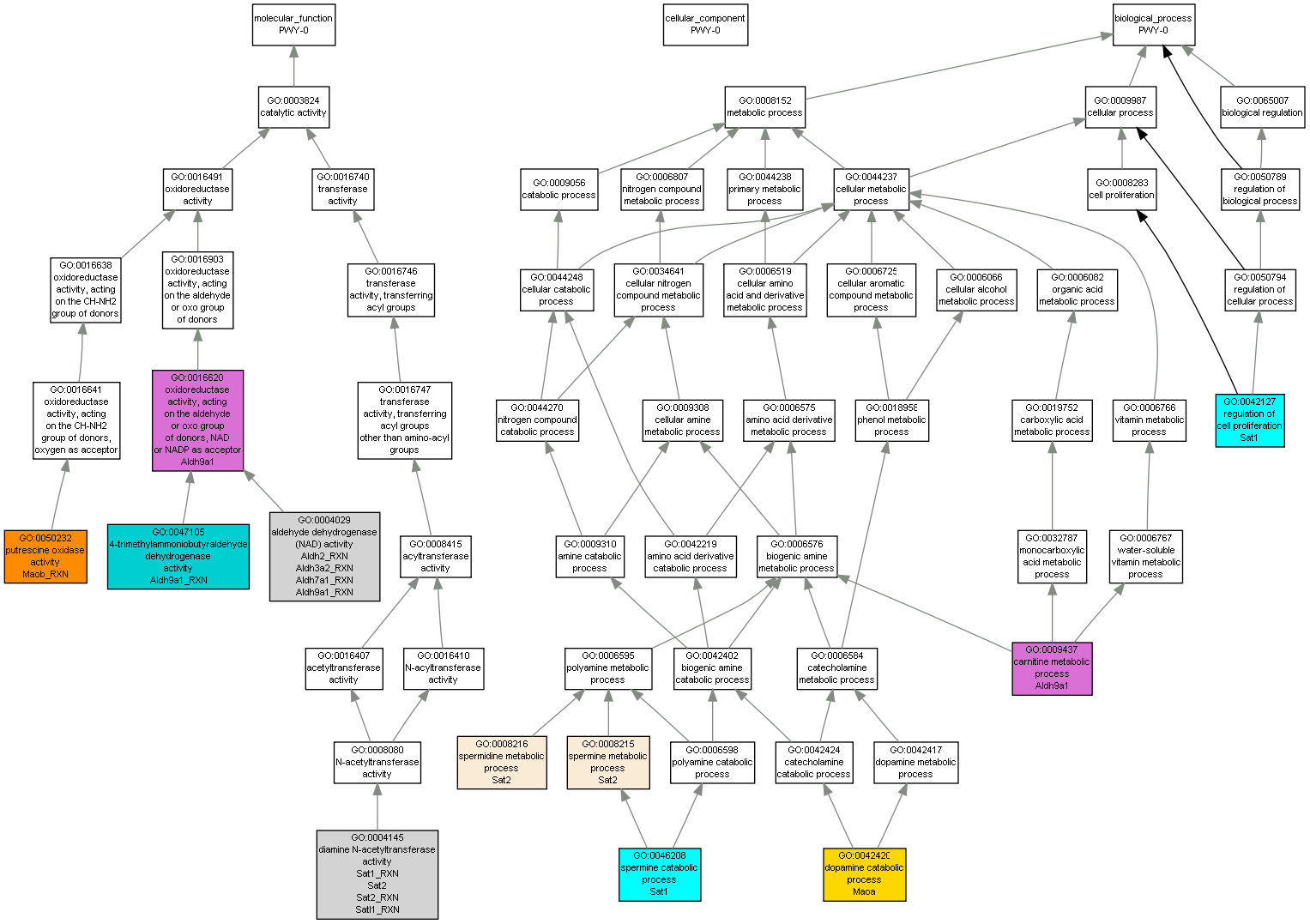
| Category | ID | Term | Mouse gene |
Evidence | Color Key |
| Molecular Function | GO:0047105 | 4-trimethylammoniobutyraldehyde dehydrogenase activity |
Aldh9a1_RXN |
MANUAL | color key |
| Molecular Function | GO:0004029 | aldehyde dehydrogenase (NAD) activity |
Aldh2_RXN |
EC-NUMBER | color key |
| Molecular Function | GO:0004029 | aldehyde dehydrogenase (NAD) activity |
Aldh3a2_RXN |
EC-NUMBER | color key |
| Molecular Function | GO:0004029 | aldehyde dehydrogenase (NAD) activity |
Aldh7a1_RXN |
EC-NUMBER | color key |
| Molecular Function | GO:0004029 | aldehyde dehydrogenase (NAD) activity |
Aldh9a1_RXN |
EC-NUMBER | color key |
| Molecular Function | GO:0004145 | diamine N-acetyltransferase activity |
Sat2 |
MGI_noIEA | color key |
| Molecular Function | GO:0004145 | diamine N-acetyltransferase activity |
Sat1_RXN |
EC-NUMBER | color key |
| Molecular Function | GO:0004145 | diamine N-acetyltransferase activity |
Sat2_RXN |
EC-NUMBER | color key |
| Molecular Function | GO:0004145 | diamine N-acetyltransferase activity |
Satl1_RXN |
EC-NUMBER | color key |
| Molecular Function | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor |
Aldh9a1 |
MGI_noIEA | color key |
| Molecular Function | GO:0050232 | putrescine oxidase activity |
Maob_RXN |
MANUAL | color key |
| Biological Process | GO:0009437 | carnitine metabolic process |
Aldh9a1 |
MGI_noIEA | color key |
| Biological Process | GO:0042420 | dopamine catabolic process |
Maoa |
MGI_noIEA | color key |
| Biological Process | GO:0042127 | regulation of cell proliferation |
Sat1 |
MGI_noIEA | color key |
| Biological Process | GO:0008216 | spermidine metabolic process |
Sat2 |
MGI_noIEA | color key |
| Biological Process | GO:0046208 | spermine catabolic process |
Sat1 |
MGI_noIEA | color key |
| Biological Process | GO:0008215 | spermine metabolic process |
Sat2 |
MGI_noIEA | color key |