|
|
|
| September 15, 2010 |
| |
- The Gene Expression Database (GXD) is pleased to announce the inclusion of RNA in situ hybridization data from the Eurexpress project. You can view the data set at accession number
J:153498. This set includes ~497,000 results for 6,594 genes in 7,052 assays with more than 118,400 images.
- Some useful links for querying subsets of data in GXD and MGI are:
- If you are interested in submitting your data to GXD, please see our guidelines for submitting expression data to the Gene Expression Database or contact gen@jax.org for further information.
|
|
| September 9, 2010 |
| |
- For the 4.4 release, MGI adds more detailed feature type categories for genes, differentiating protein coding genes from non-coding RNA genes from markers based only on heritable phenotype from immunological gene segments. You can query an entire category or limit your search to a single subcategory.
- You can query by desired feature types on the Genes and Markers Query Form. A count of the number of genes appears for each feature category.
- Feature type categories appear in the output of the Genes and Markers query and in the output from any Quick Search.
- Using MGI BioMart, you can pick a type from the MGI Feature Type list (Attributes-->Genome Features-->MGI Feature Type) and then select Feature Type as an attribute (Attributes-->Features--Feature Type). The result is a list of all MGI entities assigned that feature type.
- You can also use the MGI Batch Query Form to enter or upload a list of symbols or IDs to search on. A column identifying the feature type for each list item appears in the results.
- Many of the classification terms in MGI come from the Sequence Ontology (SO) project. The SO project develops terms and relationships describing features and attributes of biological sequences.
When the MGI classification term definition MGI comes from the SO project, the SO ID for that term appears.
-
- For details about the new categories, see Genome Feature Type Definitions.
|
|
|
| July 28, 2010 |
| |
- Genetic map position revision in MGI
- The genetic map positions for genes and markers in MGI are updated using the data and methods described in Cox et al. (2009). PubMed
- The revised standard genetic map described in Cox et al. incorporates over 10,000 single nucleotide polymorphisms (SNPs) using a set of 47 families of a heterogeneous mouse population comprising over 3500 meioses.
- The revised map corrects errors in marker order in earlier consensus genetic maps for the laboratory mouse.
- The Cox map integrates SSLP markers from other genetic maps and with physical maps of the mouse genome.
- Linear interpolation was used to translate mouse genome coordinates (NCBI Build 37) for genes and markers in MGI to sex-averaged cM locations.
- Markers in MGI that did not have genome coordinates were not updated to new cM positions; however, the original mapping data for these markers can still be found in the mapping experiment detail pages.
-
| Mapping data update summary |
|---|
| Genes and markers in MGI with interpolated cM locations:
| ~72,000 |
| Genes and markers in MGI that gained a cM position following interpolation: | ~35,000 |
| Genes and markers in MGI without an interpolated cM position (i.e. genetic map location = syntenic): | ~11,600 |
|
|
|
| June 18, 2010 |
| |
- For the 4.35 release, the MGI Quick Search returns the alleles most closely associated with your query in the Genome Features list on the results page.
As an example, type
wavy in the Quick Search box and note the various types of phenotypic allele that this query returns.
|
|
|
| May 14, 2010:
|
| |
- For the 4.34 release, MGI is retiring several query forms. See FAQ for alternative ways to find the same information on the MGI site.
|
|
| March 31, 2010:
|
| |
- MGI now represents all alleles from KOMP and EUCOMM; for example, see
Aamptm1a(KOMP)Wtsi and
Zap70tm2e(EUCOMM)Hmgu
- For the 4.33 release, gene detail pages may now include:
- A note when there are gene/pseudogene discrepancies between VEGA, Ensembl, or NCBI biotypes. Click the Biotype Conflict icon for details. As an example, see Cecr6.
A new
report lists all genes with these discrepancies.
- Vega and Ensembl transcript and/or protein sequences:
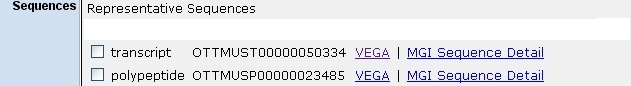
- links to gene model evidence:

- links to protein ontology annotations:
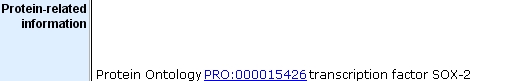
The three examples above are from the MGI Sox2 marker detail report.
- An option to direct the output of your query to Excel is now available from the following:
- Additionally, the MGI Batch Query can now handle very large files.
|
|
