|
|
|
Allele Information
|
|
Allele:
|
|
|
Synonym:
|
CYP19-Cre
|
|
Molecular description:
|
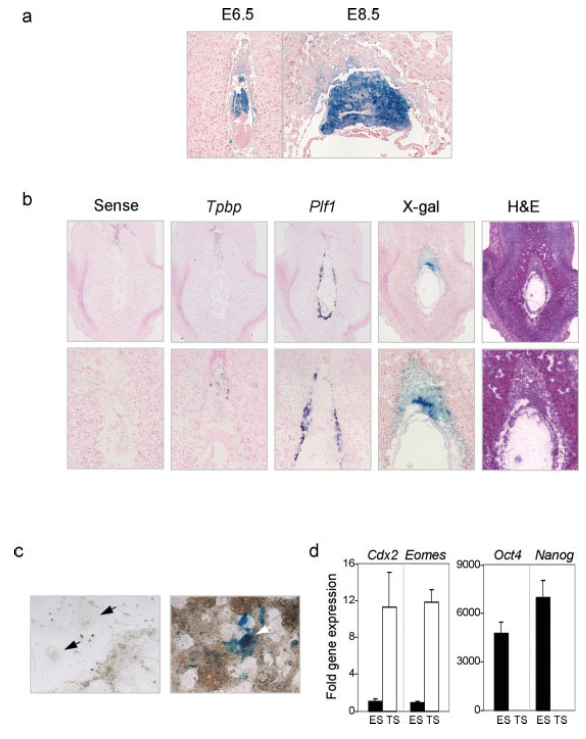
This transgene uses the cytochrome P450, family 19, subfamily a, polypeptide 1 promoter to drive expression of cre recombinase in the placenta. The transgene also contains an IRES-luciferase, the rabbit beta-globin intron and the SV-40 polyA signal. When crossed to R26R reporter mice, the 5912 founder line exhibited cre activity throughout the spongiotrophoblast and labyrinth trophoblast of E13.5 placentas (only a small percentage of fetuses display X-gal in the internal organs.) The transgene is active by E6.5 in early diploid trophoblast cells having initiated commitment to a differentiated state but is inactive in early, undifferentiated trophoblast stem cells. Strength of cre expression was related to parental inheritance, with maternal inheritance providing the strongest, most consistent placental expression. This line was studied in detail.
|
|
Find mice (IMSR):
|
Mouse Strains:
0 strains available
Cell Lines:
0 lines available
|
|
Additional Tissues:
|
Tg(Cyp19a1-cre)5912Gle activity also observed in:
embryo-other,
head,
integumental system,
nervous system,
reproductive system,
sensory organs
|
|
|
Tissue Information
|
|
Extraembryonic Component
|
Other recombinase alleles with activity in Extraembryonic Component tissues:
 Alpltm1(cre)Nagy
Alpltm1(cre)Nagy,
Cdh5em1(cre/ERT2)Smoc,
Commd10Tg(Vav1-icre)A2Kio,
Edil3Tg(Sox2-cre)1Amc,
Elf5em1(cre)Jilu,
Eomestm4.1(icre)Rob,
Esrrbtm1(cre)Yba,
Fgf18tm2.2(cre/ERT2)Dor,
Foxa2tm2.1(cre/Esr1*)Moon,
Gt(ROSA)26Sortm2(cre/ERT2)Brn,
Hlfem2(cre/ERT2)Suda,
Hnf4atm1(cre)Sdv,
Hoxa13tm1.1(cre)Mlkn,
Hoxa13tm1(cre)Mlkn,
Hprt1tm364a(CAG-icre/ERT2)Ems,
Kdrtm1(cre)Sato,
Kittm1(cre/ERT2)Dsa,
Krt19tm1(cre)Mmt,
Mapttm1(cre)Nagy,
Mecomem1(cre/ERT2)Suda,
Meox2tm1(cre)Sor,
Mesp1tm2(cre)Ysa,
Nfatc1em4(cre/ERT2)Bzsh,
Nkx1-2tm2296.1(cre/ERT2)Arte,
Nkx2-5tm2(cre)Rph,
Notch1tm3(cre)Rko,
Prdm1tm3(icre)Rob,
Prl2c2tm1(cre)Gle,
Runx1tm1(cre/Esr1*)Ims,
Runx1tm2.1(cre/Esr1*)Ims,
Shhtm2(cre/ERT2)Cjt,
Sox17tm1(icre)Heli,
Sox17tm2.1(icre)Heli,
Sox17tm4.1(icre/ERT2)Heli,
Ssttm2.1(cre)Zjh,
Synbem1(cre)Mdf,
Taglntm2(cre)Yec,
Tg(ACTA1-cre)79Jme,
Tg(Acvrl1-cre)L1Spo,
Tg(Cdh5-cre)1Spe,
Tg(Cdh5-cre)7Mlia,
Tg(Cdh5-cre/ERT2)CIVE23Mlia,
Tg(Cdx1-cre)23Kem,
Tg(Csf1r-cre/Esr1*)1Jwp,
Tg(Ddx4-cre)1Dcas,
Tg(EIIa-cre)C5379Lmgd,
Tg(Etv2-cre)15508Djg,
Tg(Etv2-cre)#Blk,
Tg(Fabp4-cre)1Rev,
Tg(Foxa3-cre)1Khk,
Tg(Gata1-cre)1Sho,
Tg(Gcm1-cre)1Chrn,
Tg(HBB-cre)12Kpe,
Tg(HBB-cre)27038Kpe,
Tg(HBB-cre)27053Kpe,
Tg(Hoxb6-cre)A5Mku,
Tg(Hoxb6-cre)C33Mku,
Tg(Hoxb6-cre)#Mku,
Tg(Hspa2-cre)1Eddy,
Tg(Itga2b-cre)NE1/21Emam,
Tg(Kdr-cre)15962Brei,
Tg(Leftb-cre)1Hmd,
Tg(Lefty1-cre/ERT2)8Hmd,
Tg(Nes-cre)1Atp,
Tg(Nrp2-cre,-EGFP)#Qsch,
Tg(Pdgfb-icre/ERT2,-EGFP)1Frut,
Tg(Prrx1-cre)1Cjt,
Tg(Sim1-cre)1Lowl,
Tg(Smarcd3-cre/ERT2)#Bbr,
Tg(Sox2-flpo)#Mhem,
Tg(Tagln-cre)1Her,
Tg(Tagln-cre)1Jomm,
Tg(Tal1-cre/ERT)1Jrg,
Tg(Tal1-cre/ERT)42-056Jrg,
Tg(T-cre)1Lwd,
Tg(Tek-cre)1Ywa,
Tg(Tek-cre)2352Rwng,
Tg(Thy1-cre)1Vln,
Tg(Tie1-cre)9Ref,
Tg(Tlx2-cre)#Ymor,
Tg(Tpbpa-cre,-EGFP)5Jcc,
Tg(Ttr-cre)1Hadj,
Tg(Vav1-cre)8Cgp,
Tg(Vil1-cre)20Syr,
Tg(Wnt11-cre/ERT2)1Jwng,
Twist2tm1.1(cre)Dor
(less)
|
|
|
Images
|
Drag images to compare to others or to data in the table below. Drag corners to resize images for more detail.
Reset Images
J:121714
Fig. 3
|
|
Recombinase Activity
|
Click heading to re-sort table.

|
|
References
|
|
 Analysis Tools
Analysis Tools




