|
|
|
Allele Information
|
|
Allele:
|
Fgf16tm1.1(cre)Sms
fibroblast growth factor 16;
targeted mutation 1.1, Suzanne L Monsour
|
Driver: Fgf16
Type: Targeted
(Recombinase)
|
|
|
Synonym:
|
Fgf16IRESCre
|
|
Molecular description:
|
Cre recombinase is expressed from this allele under control of the endogenous Fgf16 promoter in the developing cristae and spiral prominence epithelial precursors of the inner ear. The Cre recombinase coding sequence, preceded by an internal ribosomal entry site (IRES) and followed by a single frt site (marking the site of excision of a frt-flanked neomycin resistance cassette), interrupts the first exon of Fgf16 so that codon 63 is disrupted.
|
|
Find mice (IMSR):
|
Mouse Strains:
0 strains available
Cell Lines:
0 lines available
|
|
Additional Tissues:
|
Fgf16tm1.1(cre)Sms activity also observed in:
|
|
|
Tissue Information
|
|
Endocrine System
|
Other recombinase alleles with activity in Endocrine System tissues:
 7630403G23RikTg(Th-cre)1Tmd
7630403G23RikTg(Th-cre)1Tmd,
Agrptm1(cre)Lowl,
Alpltm1(cre)Nagy,
Ccktm1.1(cre)Zjh,
Cfap126tm1.1(icre)Heli,
Commd10Tg(Vav1-icre)A2Kio,
Cpa1tm1(cre/ERT2)Dam,
Crhtm1.1(flpo)Bsab,
Crhtm1(cre)Zjh,
Cx3cr1tm1.1(cre)Jung,
Cx3cr1tm2.1(cre/ERT2)Jung,
Cyp11a1tm1(GFP/cre)Pzg,
E2f1Tg(Wnt1-cre)2Sor,
Foxd1tm1(GFP/cre)Amc,
Foxl2tm1(GFP/cre/ERT2)Pzg,
Gad2tm2(cre)Zjh,
Gcgem1(cre)Utr,
Gcgtm1.1(cre/ERT2)Yasu,
Ghem1(cre)Jmz,
Ghsrtm1.1(cre)Jmz,
Glp1rtm1.1(cre)Cors,
Gnao1tm2(EGFP/cre/ERT2)Wtsi,
Golm1tm2(EGFP/cre/ERT2)Wtsi,
Gper1em1(cre)Smoc,
Gt(ROSA)26Sortm1.1(CAG-cre)Jphe,
Gt(ROSA)26Sortm1(cre/ERT2)Tyj,
Gt(ROSA)26Sortm2(icre/ERT2)Jphe,
H2az2Tg(Wnt1-cre)11Rth,
Hand2em1(dre)Pjen,
Hprt1tm1(CAG-cre)Mnn,
Hprt1tm1(TG-cre)1Sasr,
Hprt1tm364a(CAG-icre/ERT2)Ems,
Ifi208Tg(Cspg4-cre)1Akik,
Ihhem1(Tomm20/mKate2,dre)Xya,
Ins1tm1.1(cre)Thor,
Ins1tm2.1(cre/ERT2)Thor,
Krt17tm1(cre)Murr,
Krt19tm1(cre/ERT)Ggu,
Leprtm2(cre)Rck,
Lyve1tm1.1(EGFP/cre)Cys,
Mnx1tm4(cre)Tmj,
Myf5tm3(cre)Sor,
Ndor1Tg(UBC-cre/ERT2)1Ejb,
Nkx2-1tm1.1(cre/ERT2)Zjh,
Nkx2-2tm4.1(cre/EGFP)Suss,
Nkx2-5tm2(cre)Rph,
Notch1tm3(cre)Rko,
Npvftm1.1(icre)Gand,
Nxph4em1(cre)Zak,
Omptm4(cre)Mom,
Oprk1tm1.1(cre)Sros,
Osr2tm2(cre)Jian,
Oxttm1.1(icre)Ksak,
Pax3tm1(cre)Joe,
Pax7tm2.1(cre/ERT2)Fan,
Pbx3tm1(EGFP/cre/ERT2)Wtsi,
Pdx1tm1.1(dre)Gkg,
Pdyntm1.1(cre)Mjkr,
Plxnd1tm2(EGFP/cre/ERT2)Wtsi,
Prlrtm1.1(cre)Uboe,
Ptf1atm1.1(cre)Cvw,
Ptf1atm1(flpo)Hcrd,
Raxtm1.1(cre/ERT2)Sbls,
Rxfp1em1(cre)Ngai,
Setd4em1(cre)Wjy,
Setd4tm2.1(cre/ERT2)Wjy,
Sfta3tm1.1(cre/ERT2)Eem,
Shhtm1(EGFP/cre)Cjt,
Shhtm2(cre/ERT2)Cjt,
Slc17a6tm2(cre)Lowl,
Slc26a9em1(cre)Sms,
Sox2tm1.1(cre/ERT2)Jpmb,
Spink1tm3(cre)Kymm,
Sptbtm2(EGFP/cre/ERT2)Wtsi,
Ssttm2.1(cre)Zjh,
Taglntm2(cre)Yec,
Tbx3em1(cre)Mhts,
Tg(ACTA1-cre)79Jme,
Tg(ACTB-cre)1Tes,
Tg(ACTFLPe)9205Dym,
Tg(Actl6b-cre)4092Jiwu,
Tg(Adipoq-cre)1Evdr,
Tg(Akr1b7-cre)1Anm,
Tg(Alb1-cre)1Dlr,
Tg(Amh-cre)8815Reb,
Tg(Amy2-cre)1Herr,
Tg(CA2-cre)1Subw,
Tg(CA2-cre/Esr1*)1Subw,
Tg(CAG-cre/Esr1*)1Lbe,
Tg(CAG-cre/Esr1*)5Amc,
Tg(Camk2a-cre)1Nobs,
Tg(Camk2a-cre/ERT2)1Aibs,
Tg(Cdh16-cre)91Igr,
Tg(Cdh5-cre)7Mlia,
Tg(CDX2-cre)101Erf,
Tg(Cga-cre)3Sac,
Tg(Cga-icre)961Sac,
Tg(CMV-cre)1Cgn,
Tg(Crx-cre)1Tfur,
Tg(Cryaa-cre)10Mlr,
Tg(Ctgf-cre)2Aibs,
Tg(Cyp11a1-cre)16Mchu,
Tg(Cyp17a1-icre)AJako,
Tg(Cyp1a1-cre)1Dwi,
Tg(Cyp39a1-cre)1Aibs,
Tg(DBH-cre)6-8Koba,
Tg(DBH-cre)9-9Koba,
Tg(Dbh-cre/ERT2)198.1Hroh,
Tg(Ddx4-cre)1Dcas,
Tg(EIIa-cre)C5379Lmgd,
Tg(Eno2-cre)39Jme,
Tg(Fabp4-cre)1Rev,
Tg(Fev-cre)FF9Pasq,
Tg(Gcg-cre)1Herr,
Tg(Gcg-cre)1Slib,
Tg(Gcg-icre)23Cshi,
Tg(GFAP-cre)25Mes,
Tg(Gfap-cre)73.12Mvs,
Tg(Gfap-cre)77.6Mvs,
Tg(GH1-cre)1Sac,
Tg(Gh1-cre)bKnmn,
Tg(Ghrhr-cre)3242Lsk,
Tg(Ghrl-cre)E7Jmz,
Tg(Ghrl-icre)#Weiz,
Tg(Glp1r-icre)#Fmgb,
Tg(Grik4-cre)G32-4Stl,
Tg(Hoxb7-cre)13Amc,
Tg(Hspa2-cre)1Eddy,
Tg(Inha-cre)1Zuk,
Tg(Inha-cre)2Zuk,
Tg(Inha-cre)3Zuk,
Tg(Ins1-cre)25Utr,
Tg(Ins1-cre,-DsRed*T3)#Caoy,
Tg(Ins1-cre/ERT)1Lphi,
Tg(Ins1-cre/ERT2)#Pesch,
Tg(Ins2-cre)23Herr,
Tg(Ins2-cre)25Mgn,
Tg(Ins2-cre/ERT)1Dam,
Tg(Ins2-tTA,tetO-cre)1Ctlh,
Tg(INS-icre)18Msdr,
Tg(Itgax-cre)1-1Reiz,
Tg(Lck-icre)3779Nik,
Tg(Lhb-cre)1Sac,
Tg(LHX3-cre)#Sjr,
Tg(Mbp-cre)6Gvn,
Tg(Mbp-cre)9Gvn,
Tg(Mchr1-cre/ERT2)3Nbe,
Tg(MMTV-cre)4Mam,
Tg(Mx1-cre)1Cgn,
Tg(Nes-cre)1Kln,
Tg(Neurod1-cre)1Able,
Tg(Neurog3-cre)1Dam,
Tg(NEUROG3-cre)1Herr,
Tg(Neurog3-cre)C1Able,
Tg(Neurog3-cre/ERT2)1Able,
Tg(Neurog3-cre/Esr1*)1Dam,
Tg(Nkx2-1-cre)2Sand,
Tg(NPHS2-cre)295Lbh,
Tg(Nr5a1-cre)1Kmor,
Tg(Nr5a1-cre)2Klp,
Tg(Nr5a1-cre)2Lowl,
Tg(Nr5a1-cre)7Lowl,
Tg(Nr5a1-cre/ERT2)4Kmor,
Tg(Pax4-cre)1Dam,
Tg(Pax6-cre)1Rilm,
Tg(Pax6-cre,GFP)1Pgr,
Tg(Pcp2-cre)2Mpin,
Tg(Pdx1-cre)1Herr,
Tg(Pdx1-cre)6Cvw,
Tg(Pdx1-cre)89.1Dam,
Tg(Pdx1-cre/Esr1*)1Mga,
Tg(Pdx1-cre/Esr1*)35.10Dam,
Tg(Pdx1-cre/Esr1*)35.6Dam,
Tg(Pdx1-cre/Esr1*)#Dam,
Tg(Pf4-icre)Q3Rsko,
Tg(Pitx1-cre)7Rsd,
Tg(Pomc1-cre)16Lowl,
Tg(Pomc-cre)5Brn,
Tg(Pomc*-cre/ERT2)#Rlb,
Tg(Pomc*-cre)#Jdr,
Tg(Pomc-dre)#Jcbr,
Tg(Prdm1-cre)1Masu,
Tg(Prl-cre)#Salr,
Tg(Prm-cre)58Og,
Tg(Prop1-cre)1Rsd,
Tg(Prop1-icre)517Sac,
Tg(Prrx1-cre)1Cjt,
Tg(PTH-cre)4167Slib,
Tg(Rarb-cre)1Bhr,
Tg(Relb-cre)53Rie,
Tg(REN-cre)1Haca,
Tg(Ren-cre)#Kwg,
Tg(Scnn1a-cre)3Aibs,
Tg(Sim1-cre)1Lowl,
Tg(Stra8-icre)1Reb,
Tg(Sycp1-cre)4Min,
Tg(Tagln-cre)1Her,
Tg(Tek-cre)1Ywa,
Tg(tetO-cre)1Jaw,
Tg(Tg-cre)1Soff,
Tg(Tg-cre/ERT2)1Kero,
Tg(Th-cre)12Mrub,
Tg(Thy1-cre)1Vln,
Tg(TPO-cre)1Shk,
Tg(Tshb-cre)4Sac,
Tg(Tyr-cre)1Gfk,
Tg(Tyr-cre/ERT2)13Bos,
Tg(Ucp1-cre)1Evdr,
Tg(Vav1-cre)8Cgp,
Tg(Vil1-cre)997Gum,
Tg(Wfs1-cre/ERT2)3Aibs,
Thtm1(cre)Te,
Tmprss2tm1.1(cre/ERT2)Ychen,
Twist2tm1.1(cre)Dor,
Viptm1(cre)Zjh,
Wt1tm1(EGFP/cre)Wtp
(less)
|
|
|
Images
|
Drag images to compare to others or to data in the table below. Drag corners to resize images for more detail.
Reset Images
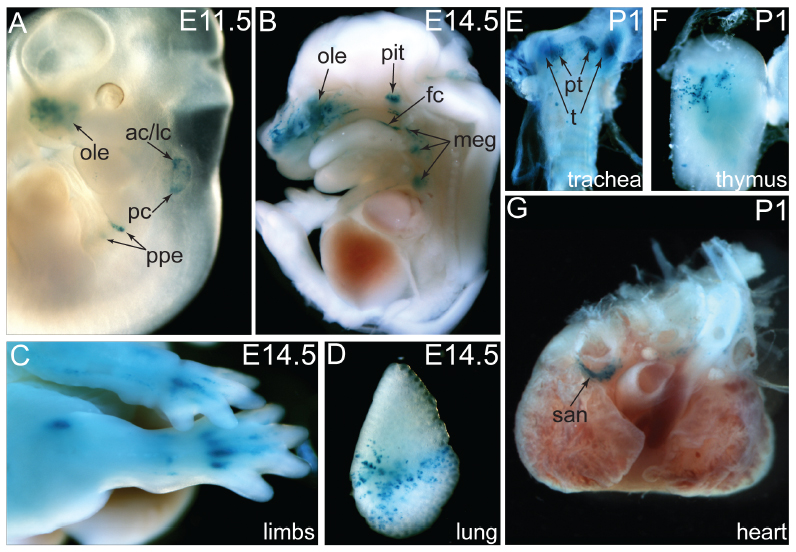
J:139718
Fig. S2
|
|
Recombinase Activity
|
Click heading to re-sort table.

|
|
References
|
|
 Analysis Tools
Analysis Tools




