|
|
|
Allele Information
|
|
Allele:
|
Tg(Scx-cre)LShuk
transgene insertion L, Chisa Shukunami
|
Driver: Scx
Type: Transgenic
(Recombinase)
|
|
|
Synonym:
|
ScxCre-L
|
|
Molecular description:
|
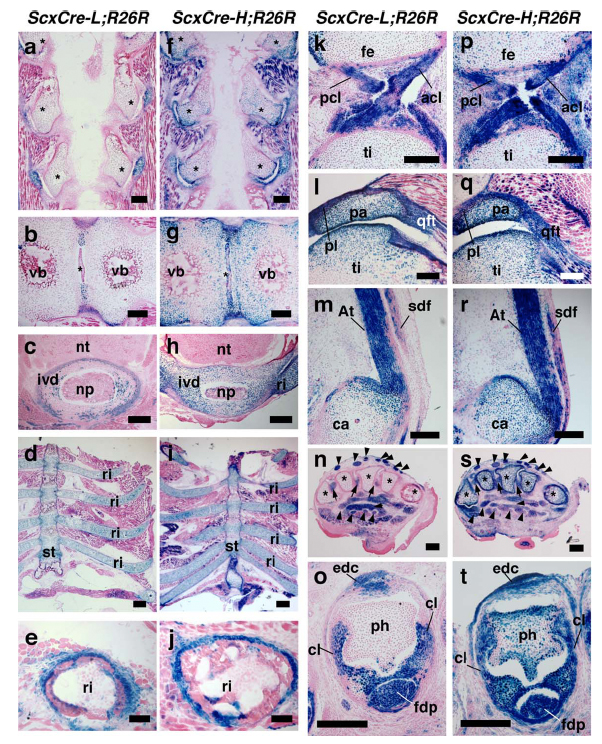
A construct was generated with the cre recombinase sequence under control of approximately 11 kb of the Scx regulatory region (~4 kb of upstream sequence and ~5 kb of downstream sequence, retrieved from a bacterial artificial chromosome RP23-415D19). This construct was used to produce transgenic animals. Two lines were established with low transgene copy number (L) and high copy number (H). When lines were bred to reporter mice, cre activity was detected overlapping various Scx-positive expression domains including chondrogenic, tendonogenic and ligamentogenic regions, as well as in the lung, kidney and testes.
|
|
Find mice (IMSR):
|
Mouse Strains:
0 strains available
Cell Lines:
0 lines available
|
|
Additional Tissues:
|
Tg(Scx-cre)LShuk activity also observed in:
limbs,
mesenchyme,
skeletal system
|
|
|
Tissue Information
|
|
Mouse-other
|
Other recombinase alleles with activity in Mouse-other tissues:
 Acantm1(cre/ERT2)Crm
Acantm1(cre/ERT2)Crm,
Alpltm1(cre)Nagy,
Amhr2em1(icre)Fjd,
Amhr2tm3(cre)Bhr,
Cd19em1.1(icre)Yoba,
Cdkn2atm1.1(cre)Dvb,
Clec9atm2.1(icre)Crs,
Cx3cr1tm1.1(cre)Jung,
Cx3cr1tm2.1(cre/ERT2)Jung,
Cxcr4tm1.1(cre/ERT2)Stum,
Dmp1tm1(cre)Msue,
Epxtm1.1(cre)Jlee,
Esrrbtm1(cre)Yba,
Fcgr1em1.1(cre)Moli,
Foxa2tm3.1(icre)Heli,
Foxp3tm4(EGFP/icre)Tch,
Foxp3tm4(YFP/icre)Ayr,
Ghem1(cre)Jmz,
Gt(ROSA)26Sorem1(CAG-Ncre/vvd*,-vvd*/Ccre)Larc,
Gt(ROSA)26Sortm13(dreo)Maoh,
Gt(ROSA)26Sortm3(CAG-Vcre)Maito,
Gt(ROSA)26Sortm4(CAG-Scre)Maito,
Hoxa11em1(cre/ERT2)Dmwe,
Hprt1tm1(CAG-cre)Mnn,
Hsd11b2tm1(cre)Anft,
Igs2tm1(Cdkn1a-cre/ERT2,-EGFP)Mxu,
Il17atm2.1(EGFP/cre/ERT2)Flv,
Krt17tm1(cre)Murr,
Lhx1tm1.1(icre)Rob,
Ly6gtm2621(cre)Arte,
Mecomem1(cre/ERT2)Suda,
Ms4a3em2(cre)Fgnx,
Ms4a3tm1(cre/ERT2)Smoc,
Nkx1-2tm2296.1(cre/ERT2)Arte,
Notch1tm3(cre)Rko,
Nox3tm1.1(cre)Kobio,
Prdm1tm3(icre)Rob,
Prdm1tm5.1(cre/ERT2)Rob,
Rbpmsem1(cre/ERT2)Yzhe,
Runx1tm1(cre/Esr1*)Ims,
Scxtm1.1(cre)Shuk,
Scxtm2.1(cre/ERT2)Stzr,
Sox17tm1(icre)Heli,
Sox17tm2.1(icre)Heli,
Tbx21tm1.1(cre/ERT2)Ayr,
Tbx21tm1.1(cre)Gmld,
Tg(Acan-cre/ERT2,-luc)1Gbg,
Tg(Acp5-cre)3Rda,
Tg(Acp5-cre)4Rda,
Tg(Acta2-cre/ERT2)1Ikal,
Tg(ACTB-cre)#Bzsh,
Tg(Acvrl1-cre)B1Spo,
Tg(Acvrl1-cre)L1Spo,
Tg(Aicda-cre,-CD2)1Hon,
Tg(Aicda-cre,-CD2)dME1Hon,
Tg(Bglap2-cre)2Kry,
Tg(CAG-cre)2Osb,
Tg(Cd4-cre)1Cwi,
Tg(Cd74-cre)#Tfk,
Tg(Cdh5-cre)1Spe,
Tg(CDX2-cre)101Erf,
Tg(Cer1-cre/ERT2)1Hmd,
Tg(Col10a1-cre)1427Vdm,
Tg(Col10a1-cre)1Xya,
Tg(Col11a2-cre)4Nhzi,
Tg(Col1a1-cre)1Bek,
Tg(Col1a1-cre)2Bek,
Tg(Col1a2-cre/ERT,-ALPP)7Cpd,
Tg(Col2a1-cre)1Bhr,
Tg(Col2a1-cre)1Star,
Tg(Col2a1-cre)1Xya,
Tg(Col2a1-cre/ERT)1Flng,
Tg(Col2a1-cre/ERT2)1Dic,
Tg(Col2a1-cre/ERT)KA3Smac,
Tg(Csrp1-cre)1Sjc,
Tg(EIIa-cre)C5379Lmgd,
Tg(Eno2-cre)39Jme,
Tg(Epx-EYFP/cre)#Voeh,
Tg(Flt3-cre)#Ccb,
Tg(Flt3-cre/ERT2)#Ccb,
Tg(Gdf5-cre,-ALPP)1Kng,
Tg(Hoxb8-dre)#Uze,
Tg(KRT14-cre)1Efu,
Tg(Lck-cre)1Cwi,
Tg(Lefty1-cre)2Hmd,
Tg(Lefty1-cre/ERT2)8Hmd,
Tg(Mcpt8-icre/ERT2)#Hkar,
Tg(Nanog-cre/Esr1*)4470Mms,
Tg(Ncr1-cre/ERT2)#Lll,
Tg(Pdgfb-icre/ERT2,-EGFP)1Frut,
Tg(Postn-cre/ERT2)#Fcko,
Tg(Prdm1-cre)1Masu,
Tg(Prg4-cre/ERT2)#Ndy,
Tg(Retnla-cre)#Voeh,
Tg(Rorc-cre)1Litt,
Tg(rtetR-tetO-cre)40Mhz,
Tg(Runx1/Hspa1a-cre/ERT2)#Itoy,
Tg(Runx2-icre)1Jtuc,
Tg(S100a4-flpo)3Lba,
Tg(Scx-cre)HShuk,
Tg(Sp7-tTA,tetO-EGFP/cre)1Amc,
Tg(Tagln-cre)1Jjl,
Tg(Tagln-cre)1Jomm,
Tg(Tbx6-cre)#Rsam,
Tg(Tek-cre)12Flv,
Tg(Tek-cre)1Ywa,
Tg(TSPY1-cre)33aYfcl,
Tg(Wnt11-cre/ERT2)1Jwng,
Tigitem1(cre/ERT2)Tebi,
Tnfsf11em2(cre)Htaka,
Tppp3tm1.1(icre/ERT2)Fan,
Tppp3tm2.1(cre/ERT2)Fan,
Twist2tm1.1(cre)Dor,
Twist2tm1(cre)Dor,
Zfp683tm1.1(cre,Hbegf)Kgis
(less)
|
|
|
Images
|
Drag images to compare to others or to data in the table below. Drag corners to resize images for more detail.
Reset Images
J:195053
Fig. 4
|
|
Recombinase Activity
|
Click heading to re-sort table.

|
|
References
|
|
 Analysis Tools
Analysis Tools




