|
|
|
Allele Information
|
|
Allele:
|
Tg(Six1-cre)2Kwk
transgene insertion 2, Kiyoshi Kawakami
|
Driver: Six1
Type: Transgenic
(Recombinase)
|
|
|
Synonym:
|
mSix1-8-NLSCre, Six1enh8-Cre, Tg(Six1-enh8-cre)2Kwk
|
|
Molecular description:
|
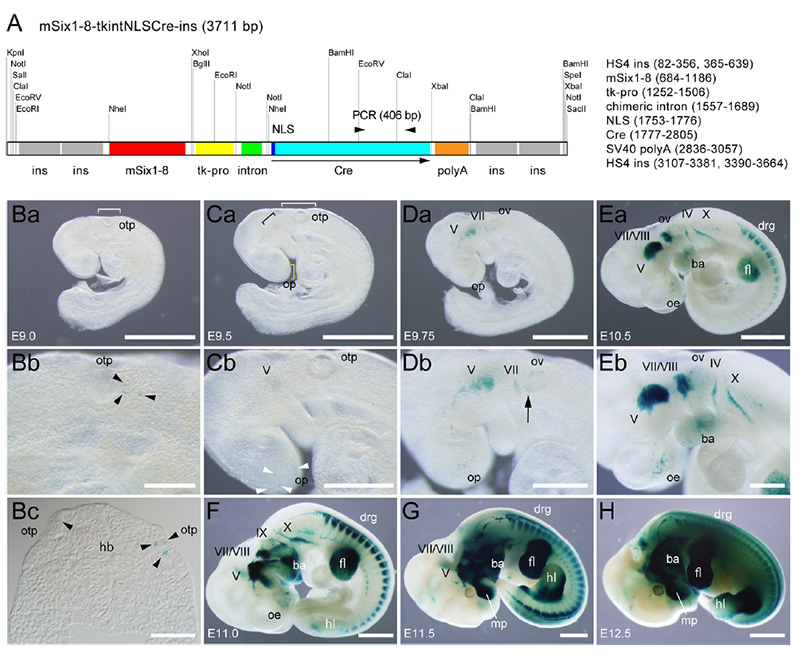
The transgene contains the sensory ganglion-specific enhancer of the Six1 homeobox gene (Six1enh8), the HSV thymidine kinase gene promoter, a chimeric intron a cre recombinase gene with a nuclear localization signal that are flanked by two tandem copies of the core region of the HS4 insulator. Lines 22L, 30L, 31L, 32L, 33L, 7l and 35l were generated. Line 35l is also designated as line 2 and was selected as the representative line.
|
|
Find mice (IMSR):
|
Mouse Strains:
0 strains available
Cell Lines:
0 lines available
|
|
Additional Tissues:
|
Tg(Six1-cre)2Kwk activity also observed in:
|
|
|
Tissue Information
|
|
Branchial Arches
|
Other recombinase alleles with activity in Branchial Arches tissues:
 7630403G23RikTg(Th-cre)1Tmd
7630403G23RikTg(Th-cre)1Tmd,
Alpltm1(cre)Nagy,
Dmbx1tm3(cre)Aoh,
E2f1Tg(Wnt1-cre)2Sor,
Emx1tm1(cre)Krj,
En1tm2(cre)Wrst,
Fgf16tm1.1(cre)Sms,
Foxa2tm1(cre)Heli,
Foxa2tm2.1(cre/Esr1*)Moon,
Foxg1tm1(cre)Skm,
Foxn1tm3(cre)Nrm,
Golm1tm2(EGFP/cre/ERT2)Wtsi,
H2az2Tg(Wnt1-cre)11Rth,
Hand2em1(dre)Pjen,
HhatTg(TFAP2A-cre)1Will,
Hoxa3tm1(cre)Moon,
Hoxb1tm1(cre)Og,
Hprt1tm364a(CAG-icre/ERT2)Ems,
Krt19tm1(cre)Mmt,
Mesp1tm2(cre)Ysa,
Myf5tm3(cre)Sor,
Nkx2-5tm1(cre)Rjs,
Nkx2-5tm2(cre)Rph,
Nkx3-2tm3.1(cre)Tlu,
Nkx3-2tm4.1(cre)Tlu,
Osr2tm2(cre)Jian,
Pax3tm1(cre)Joe,
Pax9tm1.1(cre)Sbam,
Pbx3tm1(EGFP/cre/ERT2)Wtsi,
Pdgfratm1.1(EGFP/cre/ERT2)Hyma,
Plxnd1tm2(EGFP/cre/ERT2)Wtsi,
Sema3dtm2.1(GFP/cre)Joe,
Shhtm1(EGFP/cre)Cjt,
Slc17a6tm2(cre)Lowl,
Sptbtm2(EGFP/cre/ERT2)Wtsi,
Taglntm2(cre)Yec,
Tbx1tm4(cre/Esr1*)Bld,
Tfap2atm1(cre)Moon,
Tg(ACTA1-cre)79Jme,
Tg(CAG-cre/Esr1*)5Amc,
Tg(CMV-cre)1Cgn,
Tg(Col2a1-cre)1Asz,
Tg(Ctgf-cre)2Aibs,
Tg(Cyp39a1-cre)1Aibs,
Tg(DBH-cre)6-8Koba,
Tg(DBH-cre)9-9Koba,
Tg(Ddx4-cre)1Dcas,
Tg(EIIa-cre)C5379Lmgd,
Tg(Eno2-cre)39Jme,
Tg(Fabp4-cre)1Rev,
Tgfb3tm1(cre)Vk,
Tg(Fgf15-cre)1Bld,
Tg(Fgf15/Hsp68-cre)1Bld,
Tg(Frzb-cre)1Tylz,
Tg(Gata3-cre)2476Jeng,
Tg(Gata3*-cre)2481Jeng,
Tg(GFAP-cre)25Mes,
Tg(Grik4-cre)G32-4Stl,
Tg(Hand2-cre)7-1Clou,
Tg(Hdnr-icre)5Phg,
Tg(Hspa2-cre)1Eddy,
Tg(Ins2-cre)25Mgn,
Tg(Lbx1-cre)3Cbm,
Tg(Mef2c-cre)2Blk,
Tg(mI56i-cre)6-2Clou,
Tg(Mpz-cre)94Imeg,
Tg(MYOD1-cre)M23Glh,
Tg(Nes-cre)1Atp,
Tg(NKX2-5-cre)#Kyuhl,
Tg(NKX2-5*-cre)#Kyuhl,
Tg(Pax2-cre)1Akg,
Tg(Pax3-cre)1Joe,
Tg(Pax3-cre)#Patr,
Tg(Prdm1-cre)1Masu,
Tg(Prrx1-cre)1Cjt,
Tg(Rr141-cre)1Ksec,
Tg(Sim1-cre)1Lowl,
Tg(Smad7-cre)1Sjc,
Tg(Sox10-cre)1Wdr,
Tg(Sox10-cre)507Mcln,
Tg(Sox10-ERT2/cre/ERT2)17Sor,
Tg(Sox10-icre/ERT2)388Wdr,
Tg(Tagln-cre)1Her,
Tg(Tbx18-icre)3Fech,
Tg(Tbx1-cre)1Joe,
Tg(Tek-cre)1Ywa,
Tg(Thy1-cre)1Vln,
Tg(Tyr-cre)1Gfk,
Tg(Ure2-cre)1Jlr,
Tg(Wnt11-cre/ERT2)1Jwng,
Tg(Wnt1-cre/ERT)1Alj,
Twist2tm1.1(cre)Dor,
Twist2tm1(cre)Dor,
Wt1tm1(EGFP/cre)Wtp
(less)
|
|
|
Images
|
Drag images to compare to others or to data in the table below. Drag corners to resize images for more detail.
Reset Images
J:227250
Fig. 4
|
|
Recombinase Activity
|
Click heading to re-sort table.

|
|
References
|
|
 Analysis Tools
Analysis Tools




