|
|
|
Allele Information
|
|
Allele:
|
Prdm1tm5.1(cre/ERT2)Rob
PR domain containing 1, with ZNF domain;
targeted mutation 5.1, Elizabeth J Robertson
|
Driver: Prdm1
Type: Targeted
(Inducible, Recombinase, Reporter)
|
|
|
Synonym:
|
Prdm1CreERT2, Prdm1CreERT2-LacZ
|
|
Molecular description:
|
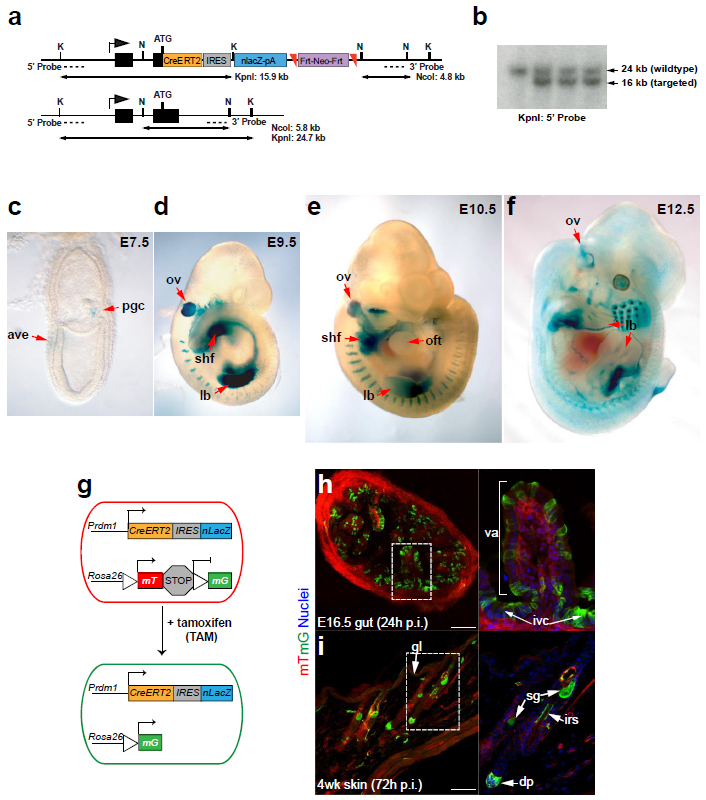
The allele was generated by introducing a cassette containing a CreERT2 upstream of IRES-nlacZ followed by a FRT-flanked neo cassette (CreERT2-IRES-nLacZ-FRT-neo-FRT) into the ATG translational site in exon 3 of the Prdm1 gene via homologous recombination in embryonic stem (ES) cells. Correctly targeted ES cell clones were transiently transfected with a FLP expression construct to excise the neo cassette.
|
|
Find mice (IMSR):
|
Mouse Strains:
0 strains available
Cell Lines:
0 lines available
|
|
Additional Tissues:
|
Prdm1tm5.1(cre/ERT2)Rob activity also observed in:
|
|
|
Tissue Information
|
|
Mouse-other
|
Other recombinase alleles with activity in Mouse-other tissues:
 Acantm1(cre/ERT2)Crm
Acantm1(cre/ERT2)Crm,
Alpltm1(cre)Nagy,
Amhr2em1(icre)Fjd,
Amhr2tm3(cre)Bhr,
Cd19em1.1(icre)Yoba,
Cdkn2atm1.1(cre)Dvb,
Clec9atm2.1(icre)Crs,
Cx3cr1tm1.1(cre)Jung,
Cx3cr1tm2.1(cre/ERT2)Jung,
Cxcr4tm1.1(cre/ERT2)Stum,
Dmp1tm1(cre)Msue,
Epxtm1.1(cre)Jlee,
Esrrbtm1(cre)Yba,
Fcgr1em1.1(cre)Moli,
Foxa2tm3.1(icre)Heli,
Foxp3tm4(EGFP/icre)Tch,
Foxp3tm4(YFP/icre)Ayr,
Ghem1(cre)Jmz,
Gt(ROSA)26Sorem1(CAG-Ncre/vvd*,-vvd*/Ccre)Larc,
Gt(ROSA)26Sortm13(dreo)Maoh,
Gt(ROSA)26Sortm3(CAG-Vcre)Maito,
Gt(ROSA)26Sortm4(CAG-Scre)Maito,
Hoxa11em1(cre/ERT2)Dmwe,
Hprt1tm1(CAG-cre)Mnn,
Hsd11b2tm1(cre)Anft,
Igs2tm1(Cdkn1a-cre/ERT2,-EGFP)Mxu,
Il17atm2.1(EGFP/cre/ERT2)Flv,
Krt17tm1(cre)Murr,
Lhx1tm1.1(icre)Rob,
Ly6gtm2621(cre)Arte,
Mecomem1(cre/ERT2)Suda,
Ms4a3em2(cre)Fgnx,
Ms4a3tm1(cre/ERT2)Smoc,
Nkx1-2tm2296.1(cre/ERT2)Arte,
Notch1tm3(cre)Rko,
Nox3tm1.1(cre)Kobio,
Prdm1tm3(icre)Rob,
Rbpmsem1(cre/ERT2)Yzhe,
Runx1tm1(cre/Esr1*)Ims,
Scxtm1.1(cre)Shuk,
Scxtm2.1(cre/ERT2)Stzr,
Sox17tm1(icre)Heli,
Sox17tm2.1(icre)Heli,
Tbx21tm1.1(cre/ERT2)Ayr,
Tbx21tm1.1(cre)Gmld,
Tg(Acan-cre/ERT2,-luc)1Gbg,
Tg(Acp5-cre)3Rda,
Tg(Acp5-cre)4Rda,
Tg(Acta2-cre/ERT2)1Ikal,
Tg(ACTB-cre)#Bzsh,
Tg(Acvrl1-cre)B1Spo,
Tg(Acvrl1-cre)L1Spo,
Tg(Aicda-cre,-CD2)1Hon,
Tg(Aicda-cre,-CD2)dME1Hon,
Tg(Bglap2-cre)2Kry,
Tg(CAG-cre)2Osb,
Tg(Cd4-cre)1Cwi,
Tg(Cd74-cre)#Tfk,
Tg(Cdh5-cre)1Spe,
Tg(CDX2-cre)101Erf,
Tg(Cer1-cre/ERT2)1Hmd,
Tg(Col10a1-cre)1427Vdm,
Tg(Col10a1-cre)1Xya,
Tg(Col11a2-cre)4Nhzi,
Tg(Col1a1-cre)1Bek,
Tg(Col1a1-cre)2Bek,
Tg(Col1a2-cre/ERT,-ALPP)7Cpd,
Tg(Col2a1-cre)1Bhr,
Tg(Col2a1-cre)1Star,
Tg(Col2a1-cre)1Xya,
Tg(Col2a1-cre/ERT)1Flng,
Tg(Col2a1-cre/ERT2)1Dic,
Tg(Col2a1-cre/ERT)KA3Smac,
Tg(Csrp1-cre)1Sjc,
Tg(EIIa-cre)C5379Lmgd,
Tg(Eno2-cre)39Jme,
Tg(Epx-EYFP/cre)#Voeh,
Tg(Flt3-cre)#Ccb,
Tg(Flt3-cre/ERT2)#Ccb,
Tg(Gdf5-cre,-ALPP)1Kng,
Tg(Hoxb8-dre)#Uze,
Tg(KRT14-cre)1Efu,
Tg(Lck-cre)1Cwi,
Tg(Lefty1-cre)2Hmd,
Tg(Lefty1-cre/ERT2)8Hmd,
Tg(Mcpt8-icre/ERT2)#Hkar,
Tg(Nanog-cre/Esr1*)4470Mms,
Tg(Ncr1-cre/ERT2)#Lll,
Tg(Pdgfb-icre/ERT2,-EGFP)1Frut,
Tg(Postn-cre/ERT2)#Fcko,
Tg(Prdm1-cre)1Masu,
Tg(Prg4-cre/ERT2)#Ndy,
Tg(Retnla-cre)#Voeh,
Tg(Rorc-cre)1Litt,
Tg(rtetR-tetO-cre)40Mhz,
Tg(Runx1/Hspa1a-cre/ERT2)#Itoy,
Tg(Runx2-icre)1Jtuc,
Tg(S100a4-flpo)3Lba,
Tg(Scx-cre)HShuk,
Tg(Scx-cre)LShuk,
Tg(Sp7-tTA,tetO-EGFP/cre)1Amc,
Tg(Tagln-cre)1Jjl,
Tg(Tagln-cre)1Jomm,
Tg(Tbx6-cre)#Rsam,
Tg(Tek-cre)12Flv,
Tg(Tek-cre)1Ywa,
Tg(TSPY1-cre)33aYfcl,
Tg(Wnt11-cre/ERT2)1Jwng,
Tigitem1(cre/ERT2)Tebi,
Tnfsf11em2(cre)Htaka,
Tppp3tm1.1(icre/ERT2)Fan,
Tppp3tm2.1(cre/ERT2)Fan,
Twist2tm1.1(cre)Dor,
Twist2tm1(cre)Dor,
Zfp683tm1.1(cre,Hbegf)Kgis
(less)
|
|
|
Images
|
Drag images to compare to others or to data in the table below. Drag corners to resize images for more detail.
Reset Images
J:257197
Fig. S1
|
|
Recombinase Activity
|
Click heading to re-sort table.

|
|
References
|
|
 Analysis Tools
Analysis Tools




