|
|
|
Allele Information
|
|
Allele:
|
|
|
Synonym:
|
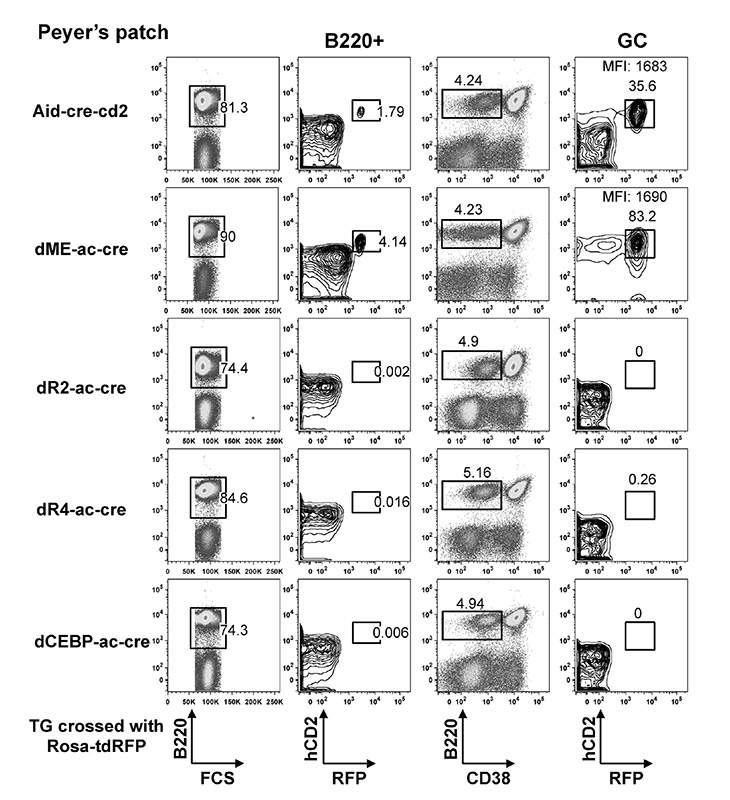
dME-ac-cre, Tg(Aicda*dME-cre,-CD2)1Hon
|
|
Molecular description:
|
The transgene contains the mouse Aicda genomic DNA with c-Myb- and E2F-binding sites (the silencing elements) that were deleted in the regulatory region 2, human CD2 cDNA reporter, Phage P1 Cre, SV40 polyA signal, yeast FRT site, and EMCV internal ribosomal entry site (ires). Two lines were generated with similar expression patterns. Line 1 is representative of all the lines.
|
|
Find mice (IMSR):
|
Mouse Strains:
0 strains available
Cell Lines:
0 lines available
|
|
Additional Tissues:
|
Tg(Aicda-cre,-CD2)dME1Hon activity also observed in:
cardiovascular system,
hemolymphoid system
|
|
|
Tissue Information
|
|
Mouse-other
|
Other recombinase alleles with activity in Mouse-other tissues:
 Acantm1(cre/ERT2)Crm
Acantm1(cre/ERT2)Crm,
Alpltm1(cre)Nagy,
Amhr2em1(icre)Fjd,
Amhr2tm3(cre)Bhr,
Cd19em1.1(icre)Yoba,
Cdkn2atm1.1(cre)Dvb,
Clec9atm2.1(icre)Crs,
Cx3cr1tm1.1(cre)Jung,
Cx3cr1tm2.1(cre/ERT2)Jung,
Cxcr4tm1.1(cre/ERT2)Stum,
Dmp1tm1(cre)Msue,
Epxtm1.1(cre)Jlee,
Esrrbtm1(cre)Yba,
Fcgr1em1.1(cre)Moli,
Foxa2tm3.1(icre)Heli,
Foxp3tm4(EGFP/icre)Tch,
Foxp3tm4(YFP/icre)Ayr,
Ghem1(cre)Jmz,
Gt(ROSA)26Sorem1(CAG-Ncre/vvd*,-vvd*/Ccre)Larc,
Gt(ROSA)26Sortm13(dreo)Maoh,
Gt(ROSA)26Sortm3(CAG-Vcre)Maito,
Gt(ROSA)26Sortm4(CAG-Scre)Maito,
Hoxa11em1(cre/ERT2)Dmwe,
Hprt1tm1(CAG-cre)Mnn,
Hsd11b2tm1(cre)Anft,
Igs2tm1(Cdkn1a-cre/ERT2,-EGFP)Mxu,
Il17atm2.1(EGFP/cre/ERT2)Flv,
Krt17tm1(cre)Murr,
Lhx1tm1.1(icre)Rob,
Ly6gtm2621(cre)Arte,
Mecomem1(cre/ERT2)Suda,
Ms4a3em2(cre)Fgnx,
Ms4a3tm1(cre/ERT2)Smoc,
Nkx1-2tm2296.1(cre/ERT2)Arte,
Notch1tm3(cre)Rko,
Nox3tm1.1(cre)Kobio,
Prdm1tm3(icre)Rob,
Prdm1tm5.1(cre/ERT2)Rob,
Rbpmsem1(cre/ERT2)Yzhe,
Runx1tm1(cre/Esr1*)Ims,
Scxtm1.1(cre)Shuk,
Scxtm2.1(cre/ERT2)Stzr,
Sox17tm1(icre)Heli,
Sox17tm2.1(icre)Heli,
Tbx21tm1.1(cre/ERT2)Ayr,
Tbx21tm1.1(cre)Gmld,
Tg(Acan-cre/ERT2,-luc)1Gbg,
Tg(Acp5-cre)3Rda,
Tg(Acp5-cre)4Rda,
Tg(Acta2-cre/ERT2)1Ikal,
Tg(ACTB-cre)#Bzsh,
Tg(Acvrl1-cre)B1Spo,
Tg(Acvrl1-cre)L1Spo,
Tg(Aicda-cre,-CD2)1Hon,
Tg(Bglap2-cre)2Kry,
Tg(CAG-cre)2Osb,
Tg(Cd4-cre)1Cwi,
Tg(Cd74-cre)#Tfk,
Tg(Cdh5-cre)1Spe,
Tg(CDX2-cre)101Erf,
Tg(Cer1-cre/ERT2)1Hmd,
Tg(Col10a1-cre)1427Vdm,
Tg(Col10a1-cre)1Xya,
Tg(Col11a2-cre)4Nhzi,
Tg(Col1a1-cre)1Bek,
Tg(Col1a1-cre)2Bek,
Tg(Col1a2-cre/ERT,-ALPP)7Cpd,
Tg(Col2a1-cre)1Bhr,
Tg(Col2a1-cre)1Star,
Tg(Col2a1-cre)1Xya,
Tg(Col2a1-cre/ERT)1Flng,
Tg(Col2a1-cre/ERT2)1Dic,
Tg(Col2a1-cre/ERT)KA3Smac,
Tg(Csrp1-cre)1Sjc,
Tg(EIIa-cre)C5379Lmgd,
Tg(Eno2-cre)39Jme,
Tg(Epx-EYFP/cre)#Voeh,
Tg(Flt3-cre)#Ccb,
Tg(Flt3-cre/ERT2)#Ccb,
Tg(Gdf5-cre,-ALPP)1Kng,
Tg(Hoxb8-dre)#Uze,
Tg(KRT14-cre)1Efu,
Tg(Lck-cre)1Cwi,
Tg(Lefty1-cre)2Hmd,
Tg(Lefty1-cre/ERT2)8Hmd,
Tg(Mcpt8-icre/ERT2)#Hkar,
Tg(Nanog-cre/Esr1*)4470Mms,
Tg(Ncr1-cre/ERT2)#Lll,
Tg(Pdgfb-icre/ERT2,-EGFP)1Frut,
Tg(Postn-cre/ERT2)#Fcko,
Tg(Prdm1-cre)1Masu,
Tg(Prg4-cre/ERT2)#Ndy,
Tg(Retnla-cre)#Voeh,
Tg(Rorc-cre)1Litt,
Tg(rtetR-tetO-cre)40Mhz,
Tg(Runx1/Hspa1a-cre/ERT2)#Itoy,
Tg(Runx2-icre)1Jtuc,
Tg(S100a4-flpo)3Lba,
Tg(Scx-cre)HShuk,
Tg(Scx-cre)LShuk,
Tg(Sp7-tTA,tetO-EGFP/cre)1Amc,
Tg(Tagln-cre)1Jjl,
Tg(Tagln-cre)1Jomm,
Tg(Tbx6-cre)#Rsam,
Tg(Tek-cre)12Flv,
Tg(Tek-cre)1Ywa,
Tg(TSPY1-cre)33aYfcl,
Tg(Wnt11-cre/ERT2)1Jwng,
Tigitem1(cre/ERT2)Tebi,
Tnfsf11em2(cre)Htaka,
Tppp3tm1.1(icre/ERT2)Fan,
Tppp3tm2.1(cre/ERT2)Fan,
Twist2tm1.1(cre)Dor,
Twist2tm1(cre)Dor,
Zfp683tm1.1(cre,Hbegf)Kgis
(less)
|
|
|
Images
|
Drag images to compare to others or to data in the table below. Drag corners to resize images for more detail.
Reset Images
J:338966
Fig. 2
|
|
Recombinase Activity
|
Click heading to re-sort table.

|
|
References
|
|
 Analysis Tools
Analysis Tools




