Gt(ROSA)26Sortm2Nat
Targeted Allele Detail
|
|
| Symbol: |
Gt(ROSA)26Sortm2Nat |
| Name: |
gene trap ROSA 26, Philippe Soriano; targeted mutation 2, Jeremy Nathans |
| MGI ID: |
MGI:3834904 |
| Synonyms: |
R26AP inv, R26IAP, ROSA26-AP inversion, Rosa26-iAP, RosaiAP |
| Gene: |
Gt(ROSA)26Sor Location: Chr6:113044389-113054205 bp, - strand Genetic Position: Chr6, 52.73 cM
|
| Alliance: |
Gt(ROSA)26Sortm2Nat page
|
|
| Germline Transmission: |
Earliest citation of germline transmission:
J:155417
|
| Parent Cell Line: |
Not Specified (ES Cell)
|
| Strain of Origin: |
129
|
|
| Allele Type: |
|
Targeted (Conditional ready, Reporter) |
| Mutation: |
|
Insertion
|
| |
|
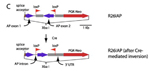
Mutation details: The human placental alkaline phosphatase (ALPP; PLAP) coding region was engineered to contain a single intron with a loxP site. Another loxP site in inverted orientation was placed 3' of the ALPP coding region. A single round of Cre-mediated recombination in E. coli resulted in the inversion of the 3' half of the coding region, thereby disrupting the ALPP open reading frame. This construct, referred to as "AP inversion" or "AP inv", was knocked into the ubiquitously-expressed Gt(ROSA)26Sor locus by homologous recombination. This allele has an inverted human placental alkaline phosphatase (ALPP; PLAP) coding segment flanked by oppositely-oriented loxP sites integrated in the Gt(ROSA)26Sor gene. Cre excision of the floxed segment flips rather than excises the ALPP inversion and reinstates a transcriptionally active form of the gene. This reporter can be activated by tissue-specific cre recombinase in any cell in the body at any time in development.
(J:155417)
|
|
Generation of the Gt(ROSA)26Sortm2Nat allele |
|
|
View phenotypes and curated references for all genotypes (concatenated display).
|
|
|
| Original: |
J:155417 Badea TC, et al., New mouse lines for the analysis of neuronal morphology using CreER(T)/loxP-directed sparse labeling. PLoS One. 2009;4(11):e7859 |
| All: |
26 reference(s) |
|
 Analysis Tools
Analysis Tools





