Zfp105Gt(TEA059)Byg
Gene trapped Allele Detail
|
|
| Symbol: |
Zfp105Gt(TEA059)Byg |
| Name: |
zinc finger protein 105; gene trap TEA059, BayGenomics |
| MGI ID: |
MGI:4128645 |
| Synonyms: |
Zfp105:LacZ, Zfp105LacZ |
| Gene: |
Zfp105 Location: Chr9:122752137-122760093 bp, + strand Genetic Position: Chr9, 73.23 cM
|
| Alliance: |
Zfp105Gt(TEA059)Byg page
|
|
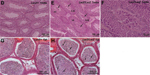
Presence of undifferentiated spermatogenic cells in the lumen of Zfp105Gt(TEA059)Byg/Zfp105Gt(TEA059)Byg testes and epididymides
Show the 1 phenotype image(s) involving this allele.
|
|
|
| Mutant Cell Line: |
TEA059 |
| Germline Transmission: |
Earliest citation of germline transmission:
J:160251
|
| Parent Cell Line: |
Other (see notes) (ES Cell)
|
| Strain of Origin: |
129P2/OlaHsd
|
|
| Allele Type: |
|
Gene trapped (Null/knockout, Reporter) |
| Mutation: |
|
Insertion of gene trap vector
Vector: pGT1Lxf
Vector Type: gene trap
|
| |
|
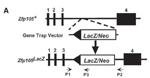
Mutation details: The gene-trap vector contains a splice-acceptor site upstream of a LacZ/Neomycin (Neo) (a fusion of LacZ and Neo). The insertion of the vector into intron 3 produces an exon 1-3-LacZ/Neo fusion protein. Northern blot analysis using radiolabeled probes against exon 4 showed the complete loss of RNA transcripts in homozygous testes. LacZ staining showed that robust expression in the testis with dominant expression in the spermatocytes (especially pachytene spermatocytes). Weak LacZ staining was also observed in a few follicles of ovaries.
(J:160251)
|
| Sequence Tags: |
|
Sequence tag details (1 tag)
| Tag ID |
GenBank ID |
Method |
Tag Location (trapped strand)* |
Select |
|
| Genome Context: |
|
Genome Browser view of this mutation
|
|
Generation of the Zfp105Gt(TEA059)Byg allele |
|
|
View phenotypes and curated references for all genotypes (concatenated display).
|
|
|
| Mouse strains and cell lines
available from the International Mouse Strain Resource
(IMSR) |
| Carrying this Mutation: |
Mouse Strains: 0 strains available
Cell Lines: 0 lines available
|
| Carrying any Zfp105 Mutation: |
21 strains or lines available
|
|
|
All BayGenomics gene trap mutations were generated in either CGR8 or E14TG2a (129P2/OlaHsd) parental ES cell lines, with the majority in subline E14TG2a.4. The specific ES cell line in which each mutation was made is not specified.
|
| Original: |
J:141210 Mouse Genome Informatics (MGI) and National Center for Biotechnology Information (NCBI), Mouse Gene Trap Data Load from dbGSS. Database Download. 2008; |
| All: |
3 reference(s) |
|
 Analysis Tools
Analysis Tools





