Prim1em1(IMPC)J
Endonuclease-mediated Allele Detail
|
|
| Symbol: |
Prim1em1(IMPC)J |
| Name: |
DNA primase, p49 subunit; endonuclease-mediated mutation 1, Jackson |
| MGI ID: |
MGI:5911622 |
| Synonyms: |
Prim1- |
| Gene: |
Prim1 Location: Chr10:127851084-127865899 bp, + strand Genetic Position: Chr10, 76.39 cM, cytoband D3
|
| Alliance: |
Prim1em1(IMPC)J page
|
| IMPC: |
Prim1 gene page |
|
Prim1em1(IMPC)J/ Prim1em1(IMPC)J mice exhibit embryonic lethality, with embryos recovered at E3.5 as blastocysts but not at E7.5. Blastocysts grown in vitro hatch from the zona pellucida and form outgrowths with apparent inner cell mass but limited/no trophectoderm or primitive endoderm cells.
Show the 1 phenotype image(s) involving this allele.
|
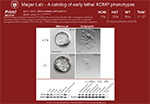
|
|
| Strain of Origin: |
C57BL/6NJ
|
| Project Collection: |
IMPC
|
|
| Allele Type: |
|
Endonuclease-mediated (Null/knockout) |
| Mutation: |
|
Intragenic deletion
|
| |
|
Mutation details: This allele was generated at The Jackson Laboratory by injecting Cas9 RNA and 4 guide sequences TAACTAAAACCTTAACTAAT, GGTTACATTATGCCCTAAAC, ACATTACCTTCAAAACTTGG and TTCTTGCCCCCAAGTTTTGA, which resulted in a 278 bp deletion beginning at Chromosome 10 negative strand position 128,016,216 bp and ending after 128,016,216 bp (GRCm38/mm10). This mutation deletes ENSMUSE00001205993 (exon 2) and 120 bp of flanking intronic sequence including the splice acceptor and donor. In addition there is a 6 bp insertion (ATCTGA) at the deletion site and a 4 bp deletion (CTAA) 33 bp after the 278 bp deletion that will not alter the results of the exon deletion. This mutation is predicted to cause a change of amino acid sequence after residue 34 and early truncation 21 amino acids later.
(J:188991)
|
| Inheritance: |
|
Not Specified |
|
|
View phenotypes and curated references for all genotypes (concatenated display).
|
|
|
|
|
| Original: |
J:188991 The Jackson Laboratory, Alleles produced for the KOMP project by The Jackson Laboratory. MGI Direct Data Submission. 2012; |
| All: |
4 reference(s) |
|
 Analysis Tools
Analysis Tools





