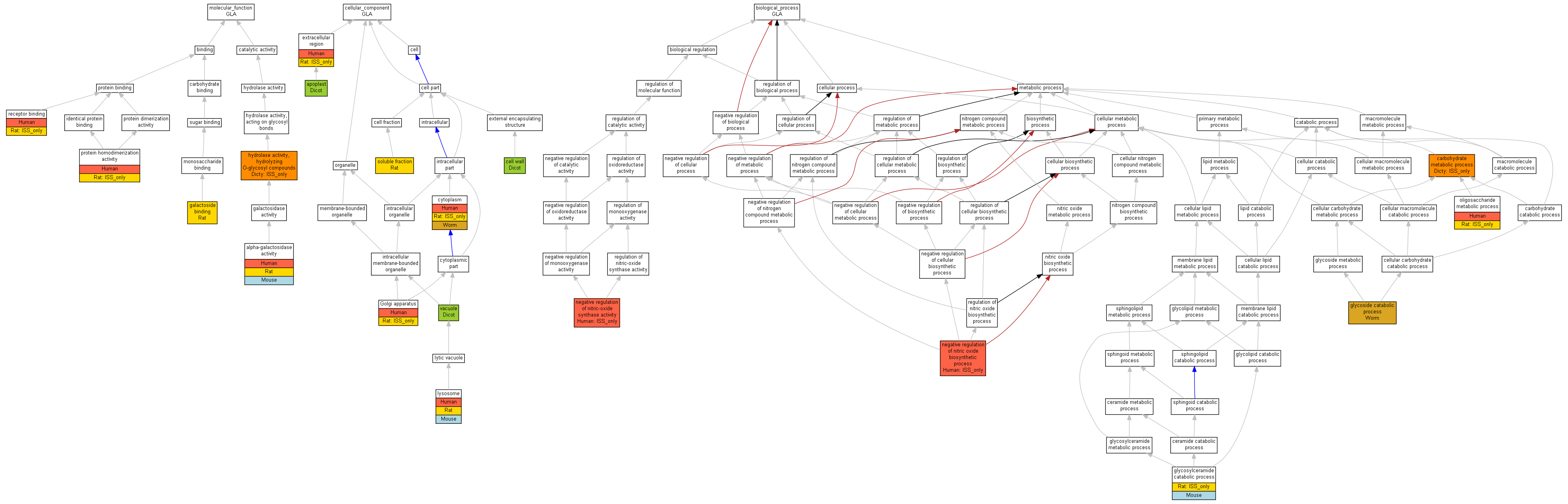
| Category | ID | Term | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005488 | binding | Human | Mouse | Rat | X | Zfish | X | Worm | Dicty | Dicot | X | X | X |
| Molecular Function | GO:0003824 | catalytic activity | Human | Mouse | Rat | X | Zfish | X | Worm | (Dicty) | Dicot | X | X | X |
| Cellular Component | GO:0005623 | cell | Human | Mouse | Rat | X | Zfish | X | Worm | Dicty | Dicot | X | X | X |
| Cellular Component | GO:0044464 | cell part | Human | Mouse | Rat | X | Zfish | X | Worm | Dicty | Dicot | X | X | X |
| Cellular Component | GO:0005576 | extracellular region | Human | Mouse | (Rat) | X | Zfish | X | Worm | Dicty | Dicot | X | X | X |
| Cellular Component | GO:0043226 | organelle | Human | Mouse | Rat | X | Zfish | X | Worm | Dicty | Dicot | X | X | X |
| Biological Process | GO:0065007 | biological regulation | (Human) | Mouse | Rat | X | Zfish | X | Worm | Dicty | Dicot | X | X | X |
| Biological Process | GO:0009987 | cellular process | (Human) | Mouse | (Rat) | X | Zfish | X | Worm | Dicty | Dicot | X | X | X |
| Biological Process | GO:0008152 | metabolic process | Human | Mouse | (Rat) | X | Zfish | X | Worm | (Dicty) | Dicot | X | X | X |
| Biological Process | GO:0048519 | negative regulation of biological process | (Human) | Mouse | Rat | X | Zfish | X | Worm | Dicty | Dicot | X | X | X |
| Biological Process | GO:0050789 | regulation of biological process | (Human) | Mouse | Rat | X | Zfish | X | Worm | Dicty | Dicot | X | X | X |
| Category | ID | Term | DB Object | Evidence | With_From | Organism | Reference |
|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0004557 | alpha-galactosidase activity | GLA | IMP | Human | PMID:16372133 | |
| Molecular Function | GO:0004557 | alpha-galactosidase activity | GLA | IDA | Human | PMID:39940 | |
| Molecular Function | GO:0004557 | alpha-galactosidase activity | Gla | IDA | Mouse | MGI:MGI:54349|PMID:198184 | |
| Molecular Function | GO:0004557 | alpha-galactosidase activity | Gla | IDA | Mouse | MGI:MGI:54860|PMID:6251472 | |
| Molecular Function | GO:0004557 | alpha-galactosidase activity | Gla | IDA | Mouse | MGI:MGI:55533|PMID:6852525 | |
| Molecular Function | GO:0004557 | alpha-galactosidase activity | Gla | IDA | Mouse | MGI:MGI:57368|PMID:2890215 | |
| Molecular Function | GO:0004557 | alpha-galactosidase activity | Gla | IDA | Mouse | MGI:MGI:58084|PMID:2906327 | |
| Molecular Function | GO:0004557 | alpha-galactosidase activity | Gla | IDA | Mouse | MGI:MGI:54440|PMID:205481 | |
| Molecular Function | GO:0004557 | alpha-galactosidase activity | Gla | IDA | Mouse | MGI:MGI:54345|PMID:895843 | |
| Molecular Function | GO:0004557 | alpha-galactosidase activity | Gla | IMP | MGI:MGI:1861794 | Mouse | MGI:MGI:86774|PMID:9122231 |
| Molecular Function | GO:0004557 | alpha-galactosidase activity | Gla | IDA | Mouse | MGI:MGI:54232|PMID:1008807 | |
| Molecular Function | GO:0004557 | alpha-galactosidase activity | Gla | IDA | Mouse | MGI:MGI:3525994|PMID:15629890 | |
| Molecular Function | GO:0004557 | alpha-galactosidase activity | Gla | IDA | Mouse | MGI:MGI:1929362|PMID:11115376 | |
| Molecular Function | GO:0004557 | alpha-galactosidase activity | Gla | IDA | Mouse | MGI:MGI:1860035|PMID:10840053 | |
| Molecular Function | GO:0004557 | alpha-galactosidase activity | Gla | ISS | RGD:10650 | Rat | RGD:1624291 |
| Molecular Function | GO:0004557 | alpha-galactosidase activity | Gla | IDA | Rat | RGD:1625498|PMID:8702598 | |
| Molecular Function | GO:0004557 | alpha-galactosidase activity | Gla | IMP | Rat | RGD:1625507|PMID:8717430 | |
| Molecular Function | GO:0004557 | alpha-galactosidase activity | Gla | ISS | RGD:1344140 | Rat | RGD:1624291 |
| Molecular Function | GO:0016936 | galactoside binding | Gla | IDA | Rat | RGD:1625498|PMID:8702598 | |
| Molecular Function | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds | DDB_G0291524 | ISS | InterPro:IPR002241 | Dicty | dictyBase_REF:10155 |
| Molecular Function | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds | melA | ISS | InterPro:IPR002241 | Dicty | dictyBase_REF:10155 |
| Molecular Function | GO:0042803 | protein homodimerization activity | GLA | IDA | Human | PMID:6256390 | |
| Molecular Function | GO:0042803 | protein homodimerization activity | Gla | ISS | RGD:1344140 | Rat | RGD:1624291 |
| Molecular Function | GO:0005102 | receptor binding | GLA | IDA | Human | PMID:1332979 | |
| Molecular Function | GO:0005102 | receptor binding | Gla | ISS | RGD:1344140 | Rat | RGD:1624291 |
| Cellular Component | GO:0048046 | apoplast | AtAGAL1 | IDA | Dicot | TAIR:Publication:501725189|PMID:18538804 | |
| Cellular Component | GO:0005618 | cell wall | AtAGAL1 | IDA | Dicot | TAIR:Publication:501714811|PMID:15593128 | |
| Cellular Component | GO:0005737 | cytoplasm | GLA | IMP | Human | PMID:1332979 | |
| Cellular Component | GO:0005737 | cytoplasm | Gla | ISS | RGD:1344140 | Rat | RGD:1624291 |
| Cellular Component | GO:0005737 | cytoplasm | GANA-1 | IDA | Worm | WB:WBPaper00024995 | |
| Cellular Component | GO:0005576 | extracellular region | GLA | IMP | Human | PMID:1332979 | |
| Cellular Component | GO:0005576 | extracellular region | GLA | IDA | Human | PMID:3029062 | |
| Cellular Component | GO:0005576 | extracellular region | Gla | ISS | RGD:1344140 | Rat | RGD:1624291 |
| Cellular Component | GO:0005794 | Golgi apparatus | GLA | IMP | Human | PMID:1332979 | |
| Cellular Component | GO:0005794 | Golgi apparatus | Gla | ISS | RGD:1344140 | Rat | RGD:1624291 |
| Cellular Component | GO:0005764 | lysosome | GLA | IMP | Human | PMID:1332979 | |
| Cellular Component | GO:0005764 | lysosome | Gla | IDA | Mouse | MGI:MGI:3525994|PMID:15629890 | |
| Cellular Component | GO:0005764 | lysosome | Gla | ISS | RGD:10650 | Rat | RGD:1624291 |
| Cellular Component | GO:0005764 | lysosome | Gla | IDA | Rat | RGD:1625498|PMID:8702598 | |
| Cellular Component | GO:0005764 | lysosome | Gla | ISS | RGD:1344140 | Rat | RGD:1624291 |
| Cellular Component | GO:0005625 | soluble fraction | Gla | IDA | Rat | RGD:1625498|PMID:8702598 | |
| Cellular Component | GO:0005773 | vacuole | AT3G56310.1 | IDA | Dicot | TAIR:Publication:501712581|PMID:15215502 | |
| Cellular Component | GO:0005773 | vacuole | AT3G56310.1 | IDA | Dicot | TAIR:Publication:501714874|PMID:15539469 | |
| Cellular Component | GO:0005773 | vacuole | AT3G56310.2 | IDA | Dicot | TAIR:Publication:501714874|PMID:15539469 | |
| Biological Process | GO:0005975 | carbohydrate metabolic process | DDB_G0291524 | ISS | InterPro:IPR002241 | Dicty | dictyBase_REF:10155 |
| Biological Process | GO:0005975 | carbohydrate metabolic process | melA | ISS | InterPro:IPR002241 | Dicty | dictyBase_REF:10155 |
| Biological Process | GO:0016139 | glycoside catabolic process | gana-1 | IMP | Worm | WB:WBPaper00024995 | |
| Biological Process | GO:0046477 | glycosylceramide catabolic process | Gla | IMP | MGI:MGI:1861794 | Mouse | MGI:MGI:86774|PMID:9122231 |
| Biological Process | GO:0046477 | glycosylceramide catabolic process | Gla | IMP | MGI:MGI:1861794 | Mouse | MGI:MGI:1859538|PMID:10841515 |
| Biological Process | GO:0046477 | glycosylceramide catabolic process | Gla | IDA | Mouse | MGI:MGI:1860035|PMID:10840053 | |
| Biological Process | GO:0046477 | glycosylceramide catabolic process | Gla | IDA | Mouse | MGI:MGI:3525994|PMID:15629890 | |
| Biological Process | GO:0046477 | glycosylceramide catabolic process | Gla | IMP | MGI:MGI:1861794 | Mouse | MGI:MGI:2155538|PMID:11752062 |
| Biological Process | GO:0046477 | glycosylceramide catabolic process | Gla | ISS | RGD:10650 | Rat | RGD:1624291 |
| Biological Process | GO:0045019 | negative regulation of nitric oxide biosynthetic process | GLA | ISS | UniProtKB:P51569 | Human | UniProtKB:P06280 |
| Biological Process | GO:0051001 | negative regulation of nitric-oxide synthase activity | GLA | ISS | UniProtKB:P51569 | Human | UniProtKB:P06280 |
| Biological Process | GO:0009311 | oligosaccharide metabolic process | GLA | IDA | Human | PMID:39940 | |
| Biological Process | GO:0009311 | oligosaccharide metabolic process | Gla | ISS | RGD:1344140 | Rat | RGD:1624291 |
| IC Inferred by curator |
| IDA Inferred from direct assay |
| IEA Inferred from electronic annotation |
| IEP Inferred from expression pattern |
| IGC Inferred from genomic context |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| ISA Inferred from sequence alignment |
| ISM Inferred from sequence model |
| ISO Inferred from sequence orthology |
| ISS Inferred from sequence or structural similarity |
| NAS Non-traceable Author Statement |
| ND No biological data available |
| RCA Reviewed computational analysis |
| TAS Traceable author statement |
| NR Not recorded (appears in some legacy annotations) |