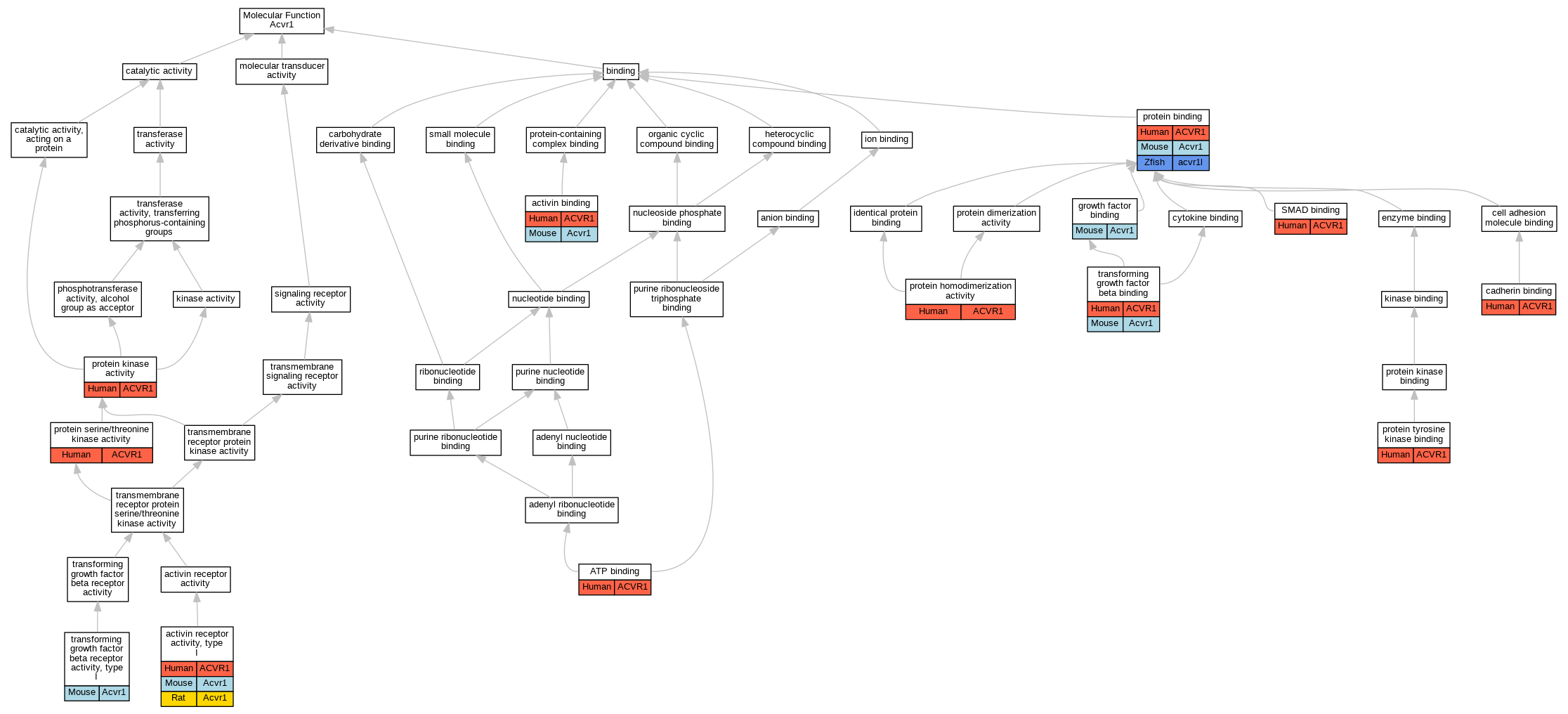
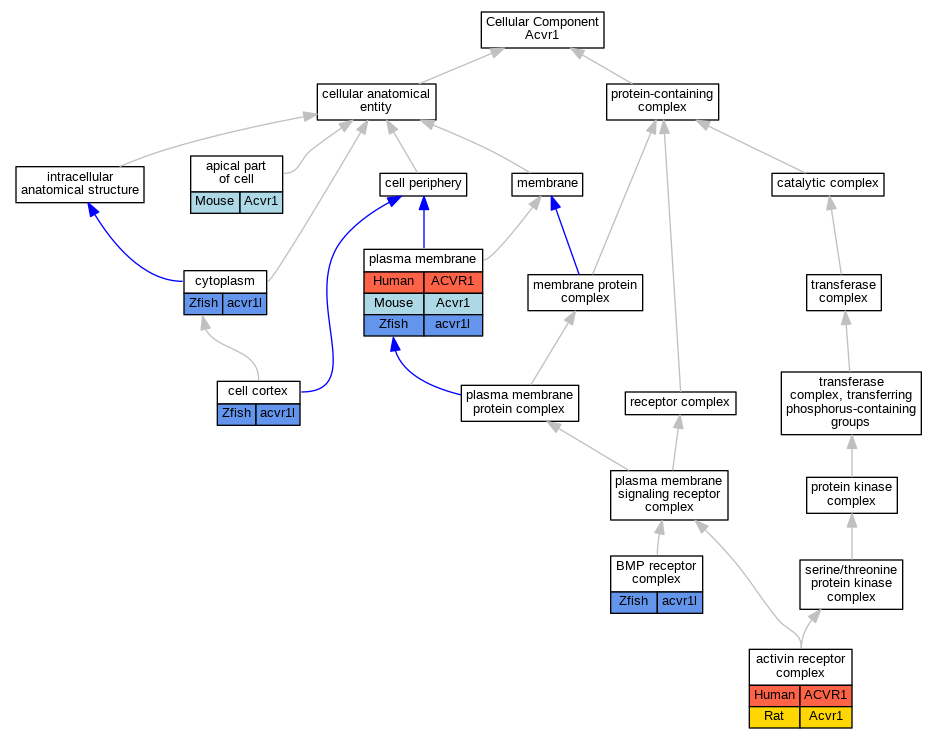
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0048185 | activin binding | ACVR1 | IDA | | Human | PMID:8242742 |
| Molecular Function | GO:0048185 | activin binding | Acvr1 | IPI | PR:P27038 | Mouse | PMID:1646080 |
| Molecular Function | GO:0048185 | activin binding | Acvr1 | IPI | PR:Q04998 | Mouse | PMID:7577669 |
| Molecular Function | GO:0016361 | activin receptor activity, type I | ACVR1 | IDA | | Human | PMID:8242742 |
| Molecular Function | GO:0016361 | activin receptor activity, type I | Acvr1 | IDA | | Mouse | PMID:11376112 |
| Molecular Function | GO:0016361 | activin receptor activity, type I | Acvr1 | IDA | | Rat | PMID:8248234|RGD:724761 |
| Molecular Function | GO:0005524 | ATP binding | ACVR1 | IDA | | Human | PMID:12065756 |
| Molecular Function | GO:0045296 | cadherin binding | ACVR1 | IPI | UniProtKB:P33151 | Human | PMID:26598555 |
| Molecular Function | GO:0019838 | growth factor binding | Acvr1 | IDA | | Mouse | PMID:8388127 |
| Molecular Function | GO:0005515 | protein binding | ACVR1 | IPI | UniProtKB:Q8N6C5 | Human | PMID:11266516 |
| Molecular Function | GO:0005515 | protein binding | ACVR1 | IPI | UniProtKB:P36894 | Human | PMID:18436533 |
| Molecular Function | GO:0005515 | protein binding | ACVR1 | IPI | UniProtKB:Q9UF33 | Human | PMID:31483918 |
| Molecular Function | GO:0005515 | protein binding | ACVR1 | IPI | UniProtKB:P27037 | Human | PMID:8242742 |
| Molecular Function | GO:0005515 | protein binding | ACVR1 | IPI | UniProtKB:P10909 | Human | PMID:8555189 |
| Molecular Function | GO:0005515 | protein binding | Acvr1 | IPI | PR:Q922H7 | Mouse | PMID:15239668 |
| Molecular Function | GO:0005515 | protein binding | acvr1l | IPI | UniProtKB:O93243 | Zfish | PMID:19377468 |
| Molecular Function | GO:0005515 | protein binding | acvr1l | IPI | ZFIN:ZDB-GENE-990415-9 | Zfish | PMID:19377468 |
| Molecular Function | GO:0042803 | protein homodimerization activity | ACVR1 | IDA | | Human | PMID:18436533 |
| Molecular Function | GO:0004672 | protein kinase activity | ACVR1 | IDA | | Human | PMID:19736306 |
| Molecular Function | GO:0004674 | protein serine/threonine kinase activity | ACVR1 | IDA | | Human | PMID:12065756 |
| Molecular Function | GO:1990782 | protein tyrosine kinase binding | ACVR1 | IPI | UniProtKB:P12931 | Human | PMID:26598555 |
| Molecular Function | GO:0046332 | SMAD binding | ACVR1 | IDA | | Human | PMID:12065756 |
| Molecular Function | GO:0050431 | transforming growth factor beta binding | ACVR1 | IDA | | Human | PMID:8242742 |
| Molecular Function | GO:0050431 | transforming growth factor beta binding | Acvr1 | IDA | | Mouse | PMID:12054694 |
| Molecular Function | GO:0005025 | transforming growth factor beta receptor activity, type I | Acvr1 | IDA | | Mouse | PMID:8388127 |
| Cellular Component | GO:0048179 | activin receptor complex | ACVR1 | IDA | | Human | PMID:8242742 |
| Cellular Component | GO:0048179 | activin receptor complex | Acvr1 | IDA | | Rat | PMID:8248234|RGD:724761 |
| Cellular Component | GO:0045177 | apical part of cell | Acvr1 | IDA | | Mouse | PMID:10704880 |
| Cellular Component | GO:0045177 | apical part of cell | Acvr1 | IDA | | Mouse | PMID:11290335 |
| Cellular Component | GO:0070724 | BMP receptor complex | acvr1l | IDA | | Zfish | PMID:19377468 |
| Cellular Component | GO:0005938 | cell cortex | acvr1l | IDA | | Zfish | PMID:18248203 |
| Cellular Component | GO:0005737 | cytoplasm | acvr1l | IDA | | Zfish | PMID:18248203 |
| Cellular Component | GO:0005886 | plasma membrane | ACVR1 | IDA | | Human | PMID:8242742 |
| Cellular Component | GO:0005886 | plasma membrane | Acvr1 | IDA | | Mouse | PMID:11376112 |
| Cellular Component | GO:0005886 | plasma membrane | acvr1l | IDA | | Zfish | PMID:32897189 |
| Biological Process | GO:0032924 | activin receptor signaling pathway | ACVR1 | IMP | | Human | PMID:18326817 |
| Biological Process | GO:0032924 | activin receptor signaling pathway | ACVR1 | IDA | | Human | PMID:19506109 |
| Biological Process | GO:0002526 | acute inflammatory response | Acvr1 | IEP | | Mouse | MGI:MGI:3693397|PMID:12151307 |
| Biological Process | GO:0009952 | anterior/posterior pattern specification | acvr1l | IMP | ZFIN:ZDB-GENO-980202-960 | Zfish | PMID:12482931 |
| Biological Process | GO:0003289 | atrial septum primum morphogenesis | ACVR1 | IMP | | Human | PMID:19506109 |
| Biological Process | GO:0003289 | atrial septum primum morphogenesis | Acvr1 | IMP | | Mouse | MGI:MGI:3610266|PMID:16140292 |
| Biological Process | GO:0003181 | atrioventricular valve morphogenesis | Acvr1 | IMP | | Mouse | MGI:MGI:3610266|PMID:16140292 |
| Biological Process | GO:0030509 | BMP signaling pathway | ACVR1 | IDA | | Human | PMID:18436533 |
| Biological Process | GO:0030509 | BMP signaling pathway | acvr1l | IMP | ZFIN:ZDB-MRPHLNO-090610-3 | Zfish | PMID:32897189 |
| Biological Process | GO:0030509 | BMP signaling pathway | acvr1l | IMP | ZFIN:ZDB-MRPHLNO-060920-2 | Zfish | PMID:32897189 |
| Biological Process | GO:0030509 | BMP signaling pathway | acvr1l | IGI | ZFIN:ZDB-GENE-000208-25 | Zfish | PMID:11222141 |
| Biological Process | GO:0030509 | BMP signaling pathway | acvr1l | IGI | ZFIN:ZDB-GENE-980526-474 | Zfish | PMID:11222141 |
| Biological Process | GO:0030509 | BMP signaling pathway | acvr1l | IGI | ZFIN:ZDB-MRPHLNO-060920-2,ZFIN:ZDB-MRPHLNO-120614-7 | Zfish | PMID:22484487 |
| Biological Process | GO:0061312 | BMP signaling pathway involved in heart development | Acvr1 | IMP | | Mouse | MGI:MGI:3610266|PMID:16140292 |
| Biological Process | GO:0007420 | brain development | acvr1l | IMP | | Zfish | PMID:14648845 |
| Biological Process | GO:0001569 | branching involved in blood vessel morphogenesis | Acvr1 | IMP | MGI:MGI:2386570,MGI:MGI:2651395 | Mouse | PMID:17078885 |
| Biological Process | GO:0003230 | cardiac atrium development | acvr1l | IMP | | Zfish | PMID:19232521 |
| Biological Process | GO:0060923 | cardiac muscle cell fate commitment | ACVR1 | IMP | | Human | PMID:19506109 |
| Biological Process | GO:0051216 | cartilage development | acvr1l | IMP | | Zfish | PMID:14648845 |
| Biological Process | GO:0071773 | cellular response to BMP stimulus | ACVR1 | IMP | | Human | PMID:18326817 |
| Biological Process | GO:0071385 | cellular response to glucocorticoid stimulus | Acvr1 | IEP | | Rat | PMID:16150914|RGD:2325236 |
| Biological Process | GO:0021556 | central nervous system formation | acvr1l | IMP | | Zfish | PMID:11165484 |
| Biological Process | GO:0061371 | determination of heart left/right asymmetry | acvr1l | IMP | ZFIN:ZDB-GENO-080731-4 | Zfish | PMID:21937597 |
| Biological Process | GO:0035462 | determination of left/right asymmetry in diencephalon | acvr1l | IMP | ZFIN:ZDB-GENO-080731-4 | Zfish | PMID:21937597 |
| Biological Process | GO:0003140 | determination of left/right asymmetry in lateral mesoderm | acvr1l | IMP | ZFIN:ZDB-GENO-080731-4 | Zfish | PMID:21937597 |
| Biological Process | GO:0007368 | determination of left/right symmetry | Acvr1 | IMP | MGI:MGI:1931734 | Mouse | PMID:15531373 |
| Biological Process | GO:0007368 | determination of left/right symmetry | acvr1l | IMP | | Zfish | PMID:9334285 |
| Biological Process | GO:0048264 | determination of ventral identity | acvr1l | IMP | | Zfish | PMID:11165484 |
| Biological Process | GO:0048264 | determination of ventral identity | acvr1l | IMP | | Zfish | PMID:11222140 |
| Biological Process | GO:0048264 | determination of ventral identity | acvr1l | IMP | ZFIN:ZDB-GENO-110804-6 | Zfish | PMID:18194657 |
| Biological Process | GO:0048264 | determination of ventral identity | acvr1l | IMP | | Zfish | PMID:12601179 |
| Biological Process | GO:0048264 | determination of ventral identity | acvr1l | IMP | ZFIN:ZDB-GENO-980202-960 | Zfish | PMID:11222141 |
| Biological Process | GO:0048264 | determination of ventral identity | acvr1l | IMP | | Zfish | PMID:9007231 |
| Biological Process | GO:0048264 | determination of ventral identity | acvr1l | IMP | | Zfish | PMID:16527746 |
| Biological Process | GO:0048264 | determination of ventral identity | acvr1l | IDA | | Zfish | PMID:11222140 |
| Biological Process | GO:0048264 | determination of ventral identity | acvr1l | IMP | ZFIN:ZDB-MRPHLNO-060920-1,ZFIN:ZDB-MRPHLNO-060920-2 | Zfish | PMID:11222140 |
| Biological Process | GO:0048264 | determination of ventral identity | acvr1l | IMP | ZFIN:ZDB-GENO-980202-960 | Zfish | PMID:11222140 |
| Biological Process | GO:0060030 | dorsal convergence | acvr1l | IMP | ZFIN:ZDB-MRPHLNO-060920-2 | Zfish | PMID:17331724 |
| Biological Process | GO:0009953 | dorsal/ventral pattern formation | acvr1l | IMP | ZFIN:ZDB-MRPHLNO-060920-2,ZFIN:ZDB-MRPHLNO-090610-3 | Zfish | PMID:19377468 |
| Biological Process | GO:0009953 | dorsal/ventral pattern formation | acvr1l | IMP | | Zfish | PMID:16775002 |
| Biological Process | GO:0009953 | dorsal/ventral pattern formation | acvr1l | IMP | | Zfish | PMID:11165484 |
| Biological Process | GO:0009953 | dorsal/ventral pattern formation | acvr1l | IGI | ZFIN:ZDB-GENE-980526-527 | Zfish | PMID:11165484 |
| Biological Process | GO:0009953 | dorsal/ventral pattern formation | acvr1l | IGI | ZFIN:ZDB-GENE-980526-373 | Zfish | PMID:12601179 |
| Biological Process | GO:0009953 | dorsal/ventral pattern formation | acvr1l | IMP | ZFIN:ZDB-GENO-980202-960 | Zfish | PMID:11222141 |
| Biological Process | GO:0009953 | dorsal/ventral pattern formation | acvr1l | IMP | ZFIN:ZDB-GENO-980202-960 | Zfish | PMID:11969259 |
| Biological Process | GO:0009953 | dorsal/ventral pattern formation | acvr1l | IMP | ZFIN:ZDB-GENO-080731-4 | Zfish | PMID:19855136 |
| Biological Process | GO:0009953 | dorsal/ventral pattern formation | acvr1l | IGI | ZFIN:ZDB-MRPHLNO-060920-2,ZFIN:ZDB-MRPHLNO-120614-7 | Zfish | PMID:22484487 |
| Biological Process | GO:0009790 | embryo development | Acvr1 | IMP | | Mouse | PMID:14729481 |
| Biological Process | GO:0009790 | embryo development | Acvr1 | IMP | MGI:MGI:2447280 | Mouse | PMID:15037318 |
| Biological Process | GO:0009790 | embryo development | Acvr1 | IMP | MGI:MGI:2651395 | Mouse | PMID:15037318 |
| Biological Process | GO:0035124 | embryonic caudal fin morphogenesis | acvr1l | IMP | ZFIN:ZDB-GENO-980202-960 | Zfish | PMID:11222141 |
| Biological Process | GO:0035050 | embryonic heart tube development | acvr1l | IMP | ZFIN:ZDB-GENO-080731-4 | Zfish | PMID:18267096 |
| Biological Process | GO:0003143 | embryonic heart tube morphogenesis | ACVR1 | IMP | | Human | PMID:19506109 |
| Biological Process | GO:0003143 | embryonic heart tube morphogenesis | acvr1l | IMP | | Zfish | PMID:9334285 |
| Biological Process | GO:0035162 | embryonic hemopoiesis | acvr1l | IMP | ZFIN:ZDB-GENO-060403-1 | Zfish | PMID:16527746 |
| Biological Process | GO:0061445 | endocardial cushion cell fate commitment | ACVR1 | IMP | | Human | PMID:19506109 |
| Biological Process | GO:0003274 | endocardial cushion fusion | Acvr1 | IMP | | Mouse | MGI:MGI:3610266|PMID:16140292 |
| Biological Process | GO:0003203 | endocardial cushion morphogenesis | Acvr1 | IMP | | Mouse | MGI:MGI:3610266|PMID:16140292 |
| Biological Process | GO:0007369 | gastrulation | Acvr1 | IMP | MGI:MGI:1857711 | Mouse | PMID:10226013 |
| Biological Process | GO:0001702 | gastrulation with mouth forming second | Acvr1 | IMP | MGI:MGI:1931734 | Mouse | PMID:17117439 |
| Biological Process | GO:0001702 | gastrulation with mouth forming second | Acvr1 | IMP | MGI:MGI:3696672 | Mouse | PMID:17117439 |
| Biological Process | GO:0007281 | germ cell development | Acvr1 | IMP | MGI:MGI:1857711 | Mouse | PMID:15289457 |
| Biological Process | GO:0007507 | heart development | Acvr1 | IMP | MGI:MGI:2386570,MGI:MGI:2651395 | Mouse | PMID:17078885 |
| Biological Process | GO:0007507 | heart development | Acvr1 | IMP | MGI:MGI:2651395 | Mouse | PMID:15226263 |
| Biological Process | GO:0007507 | heart development | Acvr1 | IMP | MGI:MGI:2136412,MGI:MGI:2651395 | Mouse | PMID:16140292 |
| Biological Process | GO:0003146 | heart jogging | acvr1l | IMP | | Zfish | PMID:9334285 |
| Biological Process | GO:0001947 | heart looping | acvr1l | IMP | | Zfish | PMID:9334285 |
| Biological Process | GO:0001947 | heart looping | acvr1l | IMP | ZFIN:ZDB-GENO-980202-960 | Zfish | PMID:17395172 |
| Biological Process | GO:0001701 | in utero embryonic development | Acvr1 | IMP | MGI:MGI:2651395 | Mouse | PMID:15037318 |
| Biological Process | GO:0001701 | in utero embryonic development | Acvr1 | IMP | | Mouse | PMID:14729481 |
| Biological Process | GO:0001701 | in utero embryonic development | Acvr1 | IMP | MGI:MGI:3696672 | Mouse | PMID:17117439 |
| Biological Process | GO:0001701 | in utero embryonic development | Acvr1 | IMP | MGI:MGI:2447280 | Mouse | PMID:15037318 |
| Biological Process | GO:0001701 | in utero embryonic development | Acvr1 | IMP | MGI:MGI:1931734 | Mouse | PMID:17117439 |
| Biological Process | GO:0048368 | lateral mesoderm development | acvr1l | IMP | | Zfish | PMID:19232521 |
| Biological Process | GO:0001889 | liver development | acvr1l | IMP | ZFIN:ZDB-GENO-980202-960 | Zfish | PMID:17507405 |
| Biological Process | GO:0007498 | mesoderm development | Acvr1 | IMP | MGI:MGI:1931734 | Mouse | PMID:17117439 |
| Biological Process | GO:0007498 | mesoderm development | Acvr1 | IMP | MGI:MGI:3696672 | Mouse | PMID:17117439 |
| Biological Process | GO:0001707 | mesoderm formation | Acvr1 | IMP | MGI:MGI:1931734 | Mouse | PMID:17117439 |
| Biological Process | GO:0001707 | mesoderm formation | Acvr1 | IMP | MGI:MGI:1931734 | Mouse | PMID:10479450 |
| Biological Process | GO:0001707 | mesoderm formation | Acvr1 | IMP | MGI:MGI:3696672 | Mouse | PMID:17117439 |
| Biological Process | GO:0048332 | mesoderm morphogenesis | acvr1l | IMP | | Zfish | PMID:11165484 |
| Biological Process | GO:0003183 | mitral valve morphogenesis | ACVR1 | IMP | | Human | PMID:19506109 |
| Biological Process | GO:0032926 | negative regulation of activin receptor signaling pathway | ACVR1 | IMP | | Human | PMID:9884026 |
| Biological Process | GO:0046588 | negative regulation of calcium-dependent cell-cell adhesion | acvr1l | IDA | | Zfish | PMID:17331724 |
| Biological Process | GO:0046588 | negative regulation of calcium-dependent cell-cell adhesion | acvr1l | IMP | ZFIN:ZDB-MRPHLNO-060920-2 | Zfish | PMID:17331724 |
| Biological Process | GO:2001237 | negative regulation of extrinsic apoptotic signaling pathway | ACVR1 | IMP | | Human | PMID:9884026 |
| Biological Process | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle | ACVR1 | IMP | | Human | PMID:9884026 |
| Biological Process | GO:0009968 | negative regulation of signal transduction | ACVR1 | IMP | | Human | PMID:9884026 |
| Biological Process | GO:0001755 | neural crest cell migration | Acvr1 | IMP | MGI:MGI:2651395 | Mouse | PMID:15226263 |
| Biological Process | GO:0042476 | odontogenesis | acvr1l | IEP | | Zfish | PMID:14745232 |
| Biological Process | GO:0042476 | odontogenesis | acvr1l | IMP | | Zfish | PMID:11759004 |
| Biological Process | GO:0001503 | ossification | acvr1l | IMP | ZFIN:ZDB-GENO-190109-3 | Zfish | PMID:28394244 |
| Biological Process | GO:0060389 | pathway-restricted SMAD protein phosphorylation | ACVR1 | IDA | | Human | PMID:19736306 |
| Biological Process | GO:0018107 | peptidyl-threonine phosphorylation | ACVR1 | IDA | | Human | PMID:19736306 |
| Biological Process | GO:0060037 | pharyngeal system development | Acvr1 | IMP | MGI:MGI:2386570,MGI:MGI:2651395 | Mouse | PMID:17078885 |
| Biological Process | GO:0030501 | positive regulation of bone mineralization | ACVR1 | IMP | | Human | PMID:18436533 |
| Biological Process | GO:0030335 | positive regulation of cell migration | ACVR1 | IGI | UniProtKB:P17813 | Human | PMID:19736306 |
| Biological Process | GO:2000017 | positive regulation of determination of dorsal identity | ACVR1 | IDA | | Human | PMID:19506109 |
| Biological Process | GO:0045893 | positive regulation of DNA-templated transcription | ACVR1 | IDA | | Human | PMID:8242742 |
| Biological Process | GO:1905007 | positive regulation of epithelial to mesenchymal transition involved in endocardial cushion formation | Acvr1 | IMP | | Mouse | MGI:MGI:3610266|PMID:16140292 |
| Biological Process | GO:0045669 | positive regulation of osteoblast differentiation | ACVR1 | IMP | | Human | PMID:18436533 |
| Biological Process | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation | ACVR1 | IDA | | Human | PMID:19506109 |
| Biological Process | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation | Acvr1 | IMP | | Mouse | MGI:MGI:3610266|PMID:16140292 |
| Biological Process | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation | ACVR1 | IMP | | Human | PMID:26598555 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | ACVR1 | IMP | | Human | PMID:18326817 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | ACVR1 | IDA | | Human | PMID:19506109 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | Acvr1 | IMP | | Mouse | MGI:MGI:3610266|PMID:16140292 |
| Biological Process | GO:0003342 | proepicardium development | acvr1l | IMP | ZFIN:ZDB-GENO-080731-4 | Zfish | PMID:20413782 |
| Biological Process | GO:0006468 | protein phosphorylation | ACVR1 | IDA | | Human | PMID:12065756 |
| Biological Process | GO:0006468 | protein phosphorylation | ACVR1 | IDA | | Human | PMID:19506109 |
| Biological Process | GO:0030510 | regulation of BMP signaling pathway | acvr1l | IMP | ZFIN:ZDB-GENO-980202-960 | Zfish | PMID:14648845 |
| Biological Process | GO:0030510 | regulation of BMP signaling pathway | acvr1l | IGI | ZFIN:ZDB-GENE-980526-373 | Zfish | PMID:12601179 |
| Biological Process | GO:0030278 | regulation of ossification | ACVR1 | IMP | | Human | PMID:16642017 |
| Biological Process | GO:0048641 | regulation of skeletal muscle tissue development | Acvr1 | IEP | | Rat | PMID:12968668|RGD:8554636 |
| Biological Process | GO:0051145 | smooth muscle cell differentiation | Acvr1 | IMP | MGI:MGI:2386570,MGI:MGI:2651395 | Mouse | PMID:17078885 |
| Biological Process | GO:0007179 | transforming growth factor beta receptor signaling pathway | ACVR1 | IDA | | Human | PMID:8242742 |
| Biological Process | GO:0007179 | transforming growth factor beta receptor signaling pathway | Acvr1 | IDA | | Mouse | PMID:11376112 |
| Biological Process | GO:0007179 | transforming growth factor beta receptor signaling pathway | Acvr1 | IMP | | Rat | PMID:12968668|RGD:8554636 |
| Biological Process | GO:0001655 | urogenital system development | Acvr1 | IMP | | Rat | PMID:11376112|RGD:8554753 |
| Biological Process | GO:0001944 | vasculature development | acvr1l | IMP | ZFIN:ZDB-GENO-980202-960 | Zfish | PMID:11222141 |
| Biological Process | GO:0060412 | ventricular septum morphogenesis | Acvr1 | IMP | | Mouse | MGI:MGI:3610266|PMID:16140292 |
 Analysis Tools
Analysis Tools