|
Source |
Alliance of Genome Resources |
| Category | ID | Classification term | Gene | Evidence | Inferred from | Organism | Reference |
|---|---|---|---|---|---|---|---|
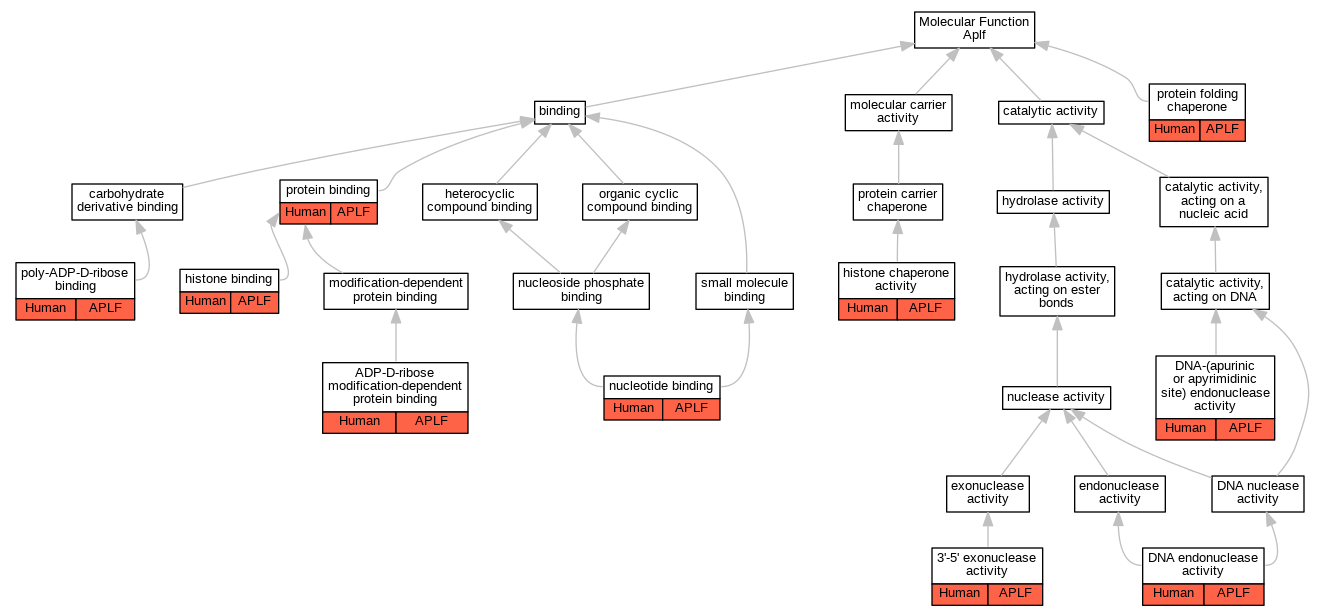
| Molecular Function | GO:0008408 | 3'-5' exonuclease activity | APLF | IDA | Human | PMID:17396150 | |
| Molecular Function | GO:0160002 | ADP-D-ribose modification-dependent protein binding | APLF | IDA | Human | PMID:18172500 | |
| Molecular Function | GO:0004520 | DNA endonuclease activity | APLF | IDA | Human | PMID:17396150 | |
| Molecular Function | GO:0003906 | DNA-(apurinic or apyrimidinic site) endonuclease activity | APLF | IDA | Human | PMID:17396150 | |
| Molecular Function | GO:0042393 | histone binding | APLF | IDA | Human | PMID:21211722 | |
| Molecular Function | GO:0042393 | histone binding | APLF | IDA | Human | PMID:29905837 | |
| Molecular Function | GO:0140713 | histone chaperone activity | APLF | IDA | Human | PMID:21211722 | |
| Molecular Function | GO:0140713 | histone chaperone activity | APLF | IDA | Human | PMID:29905837 | |
| Molecular Function | GO:0000166 | nucleotide binding | APLF | IDA | Human | PMID:18172500 | |
| Molecular Function | GO:0072572 | poly-ADP-D-ribose binding | APLF | IDA | Human | PMID:18172500 | |
| Molecular Function | GO:0072572 | poly-ADP-D-ribose binding | APLF | IDA | Human | PMID:21211721 | |
| Molecular Function | GO:0072572 | poly-ADP-D-ribose binding | APLF | IDA | Human | PMID:30104678 | |
| Molecular Function | GO:0005515 | protein binding | APLF | IPI | UniProtKB:P18887 | Human | PMID:17353262 |
| Molecular Function | GO:0005515 | protein binding | APLF | IPI | UniProtKB:Q13426 | Human | PMID:17353262 |
| Molecular Function | GO:0005515 | protein binding | APLF | IPI | UniProtKB:P09874 | Human | PMID:17396150 |
| Molecular Function | GO:0005515 | protein binding | APLF | IPI | UniProtKB:P13010 | Human | PMID:17396150 |
| Molecular Function | GO:0005515 | protein binding | APLF | IPI | UniProtKB:P18887 | Human | PMID:17396150 |
| Molecular Function | GO:0005515 | protein binding | APLF | IPI | UniProtKB:P49917 | Human | PMID:17396150 |
| Molecular Function | GO:0005515 | protein binding | APLF | IPI | UniProtKB:Q13426 | Human | PMID:17396150 |
| Molecular Function | GO:0005515 | protein binding | APLF | IPI | UniProtKB:P09874 | Human | PMID:18172500 |
| Molecular Function | GO:0005515 | protein binding | APLF | IPI | UniProtKB:P09874 | Human | PMID:20098424 |
| Molecular Function | GO:0005515 | protein binding | APLF | IPI | UniProtKB:P13010 | Human | PMID:20098424 |
| Molecular Function | GO:0005515 | protein binding | APLF | IPI | UniProtKB:P13010 | Human | PMID:23178593 |
| Molecular Function | GO:0005515 | protein binding | APLF | IPI | UniProtKB:P18887 | Human | PMID:23178593 |
| Molecular Function | GO:0005515 | protein binding | APLF | IPI | UniProtKB:P49917 | Human | PMID:23178593 |
| Molecular Function | GO:0005515 | protein binding | APLF | IPI | UniProtKB:Q13426 | Human | PMID:23178593 |
| Molecular Function | GO:0005515 | protein binding | APLF | IPI | UniProtKB:P12956 | Human | PMID:23689425 |
| Molecular Function | GO:0005515 | protein binding | APLF | IPI | UniProtKB:P13010 | Human | PMID:23689425 |
| Molecular Function | GO:0005515 | protein binding | APLF | IPI | UniProtKB:Q53SE7 | Human | PMID:25416956 |
| Molecular Function | GO:0005515 | protein binding | APLF | IPI | UniProtKB:P09874 | Human | PMID:28514442 |
| Molecular Function | GO:0005515 | protein binding | APLF | IPI | UniProtKB:P13010 | Human | PMID:28514442 |
| Molecular Function | GO:0005515 | protein binding | APLF | IPI | UniProtKB:P18887 | Human | PMID:28514442 |
| Molecular Function | GO:0005515 | protein binding | APLF | IPI | UniProtKB:P49917 | Human | PMID:28514442 |
| Molecular Function | GO:0005515 | protein binding | APLF | IPI | UniProtKB:Q13426 | Human | PMID:28514442 |
| Molecular Function | GO:0044183 | protein folding chaperone | APLF | EXP | Human | PMID:29905837 | |
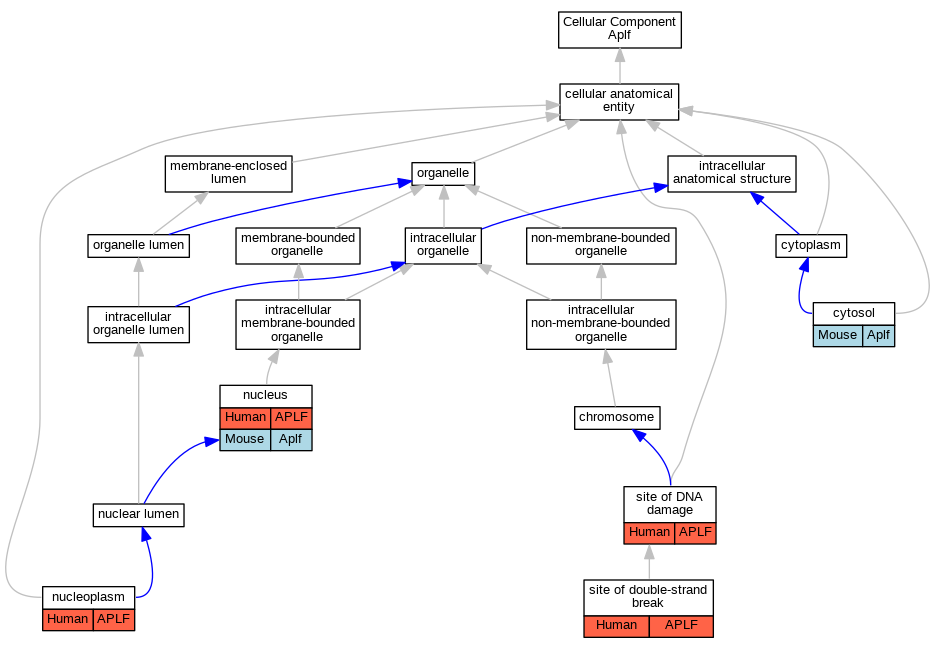
| Cellular Component | GO:0005829 | cytosol | Aplf | IDA | Mouse | MGI:MGI:6780835|PMID:33277378 | |
| Cellular Component | GO:0005654 | nucleoplasm | APLF | IDA | Human | GO_REF:0000052 | |
| Cellular Component | GO:0005634 | nucleus | APLF | IDA | Human | PMID:17396150 | |
| Cellular Component | GO:0005634 | nucleus | APLF | IDA | Human | PMID:18172500 | |
| Cellular Component | GO:0005634 | nucleus | Aplf | IDA | Mouse | MGI:MGI:6780835|PMID:33277378 | |
| Cellular Component | GO:0090734 | site of DNA damage | APLF | IDA | Human | PMID:30104678 | |
| Cellular Component | GO:0090734 | site of DNA damage | APLF | IDA | Human | PMID:21211722 | |
| Cellular Component | GO:0035861 | site of double-strand break | APLF | IDA | Human | PMID:23689425 | |
| Cellular Component | GO:0035861 | site of double-strand break | APLF | IDA | Human | PMID:21211721 | |
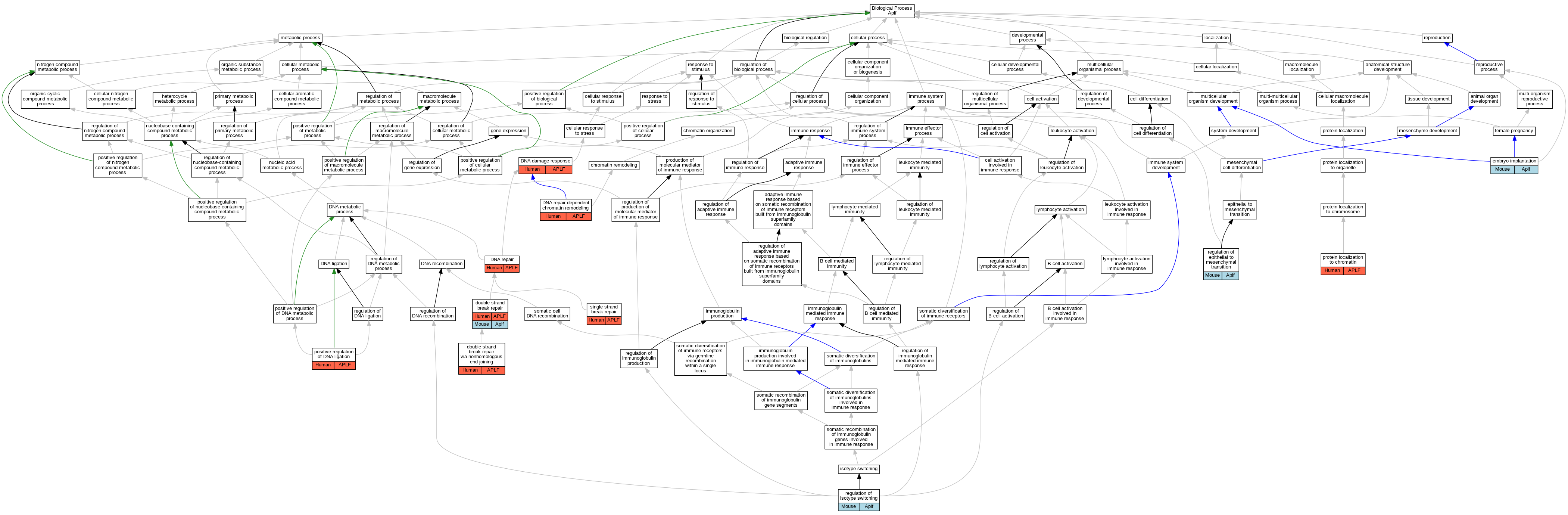
| Biological Process | GO:0006974 | DNA damage response | APLF | IDA | Human | PMID:17396150 | |
| Biological Process | GO:0006281 | DNA repair | APLF | IDA | Human | PMID:21211722 | |
| Biological Process | GO:0006281 | DNA repair | APLF | IDA | Human | PMID:30104678 | |
| Biological Process | GO:0140861 | DNA repair-dependent chromatin remodeling | APLF | IDA | Human | PMID:21211722 | |
| Biological Process | GO:0140861 | DNA repair-dependent chromatin remodeling | APLF | IDA | Human | PMID:29905837 | |
| Biological Process | GO:0006302 | double-strand break repair | APLF | IMP | Human | PMID:17396150 | |
| Biological Process | GO:0006302 | double-strand break repair | Aplf | IMP | MGI:MGI:4128102 | Mouse | PMID:21211721 |
| Biological Process | GO:0006303 | double-strand break repair via nonhomologous end joining | APLF | IMP | Human | PMID:23689425 | |
| Biological Process | GO:0007566 | embryo implantation | Aplf | IMP | Mouse | MGI:MGI:6780835|PMID:33277378 | |
| Biological Process | GO:0051106 | positive regulation of DNA ligation | APLF | IGI | UniProtKB:Q9Y6F1 | Human | PMID:21211721 |
| Biological Process | GO:0071168 | protein localization to chromatin | APLF | IDA | Human | PMID:21211722 | |
| Biological Process | GO:0010717 | regulation of epithelial to mesenchymal transition | Aplf | IMP | Mouse | MGI:MGI:6780835|PMID:33277378 | |
| Biological Process | GO:0045191 | regulation of isotype switching | Aplf | IMP | MGI:MGI:4128102 | Mouse | PMID:21211721 |
| Biological Process | GO:0000012 | single strand break repair | APLF | IMP | Human | PMID:17396150 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 06/12/2024 MGI 6.13 |

|
|
|
||