|
Source |
Alliance of Genome Resources |
| Category | ID | Classification term | Gene | Evidence | Inferred from | Organism | Reference |
|---|---|---|---|---|---|---|---|
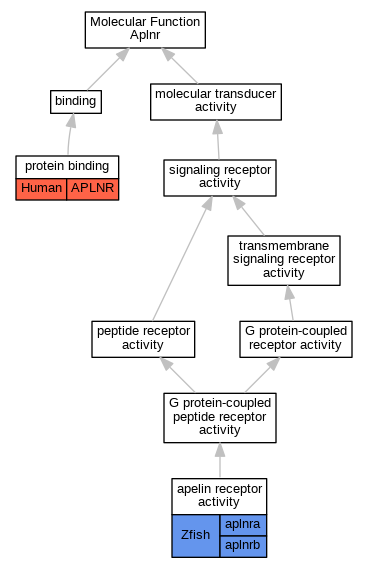
| Molecular Function | GO:0060182 | apelin receptor activity | aplnra | IDA | Zfish | PMID:30631305 | |
| Molecular Function | GO:0060182 | apelin receptor activity | aplnrb | IDA | Zfish | PMID:30631305 | |
| Molecular Function | GO:0060182 | apelin receptor activity | aplnrb | IDA | Zfish | PMID:17336906 | |
| Molecular Function | GO:0005515 | protein binding | APLNR | IPI | UniProtKB:P30556 | Human | PMID:21412239 |
| Molecular Function | GO:0005515 | protein binding | APLNR | IPI | UniProtKB:P41145 | Human | PMID:22200678 |
| Molecular Function | GO:0005515 | protein binding | APLNR | IPI | UniProtKB:P30556 | Human | PMID:22935142 |
| Molecular Function | GO:0005515 | protein binding | APLNR | IPI | UniProtKB:P29033 | Human | PMID:32296183 |
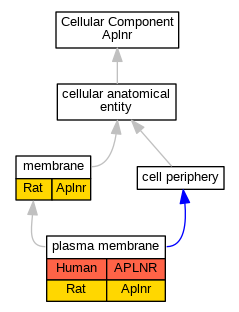
| Cellular Component | GO:0016020 | membrane | Aplnr | IDA | Rat | PMID:14645236|RGD:1304466 | |
| Cellular Component | GO:0005886 | plasma membrane | APLNR | IDA | Human | PMID:25639753 | |
| Cellular Component | GO:0005886 | plasma membrane | Aplnr | IDA | Rat | PMID:11359874|RGD:12907555 | |
| Biological Process | GO:0007512 | adult heart development | Aplnr | IMP | Mouse | MGI:MGI:5913613|PMID:28663440 | |
| Biological Process | GO:0035479 | angioblast cell migration from lateral mesoderm to midline | aplnrb | IGI | ZFIN:ZDB-GENE-060929-512 | Zfish | PMID:26017639 |
| Biological Process | GO:0035479 | angioblast cell migration from lateral mesoderm to midline | aplnrb | IMP | ZFIN:ZDB-MRPHLNO-070522-4 | Zfish | PMID:26017639 |
| Biological Process | GO:0035479 | angioblast cell migration from lateral mesoderm to midline | aplnra | IGI | ZFIN:ZDB-GENE-050913-90 | Zfish | PMID:26017639 |
| Biological Process | GO:0035479 | angioblast cell migration from lateral mesoderm to midline | aplnra | IMP | ZFIN:ZDB-MRPHLNO-070525-6 | Zfish | PMID:26017639 |
| Biological Process | GO:0035904 | aorta development | Aplnr | IMP | Mouse | MGI:MGI:5584055|PMID:23603510 | |
| Biological Process | GO:0060183 | apelin receptor signaling pathway | Aplnr | IMP | Mouse | MGI:MGI:5584055|PMID:23603510 | |
| Biological Process | GO:0060183 | apelin receptor signaling pathway | aplnrb | IDA | Zfish | PMID:30631305|ZFIN:ZDB-PUB-190113-16 | |
| Biological Process | GO:0003171 | atrioventricular valve development | Aplnr | IMP | Mouse | MGI:MGI:5584055|PMID:23603510 | |
| Biological Process | GO:0048738 | cardiac muscle tissue development | aplnrb | IMP | Zfish | PMID:17336906|ZFIN:ZDB-PUB-070310-9 | |
| Biological Process | GO:0016477 | cell migration | aplnrb | IMP | Zfish | PMID:17336906|ZFIN:ZDB-PUB-070310-9 | |
| Biological Process | GO:0042074 | cell migration involved in gastrulation | aplnra | IMP | ZFIN:ZDB-MRPHLNO-110413-7 | Zfish | PMID:19922670 |
| Biological Process | GO:0042074 | cell migration involved in gastrulation | aplnra | IMP | ZFIN:ZDB-MRPHLNO-110413-2 | Zfish | PMID:19922670 |
| Biological Process | GO:0042074 | cell migration involved in gastrulation | aplnra | IMP | ZFIN:ZDB-MRPHLNO-110413-3 | Zfish | PMID:19922670 |
| Biological Process | GO:0060976 | coronary vasculature development | Aplnr | IMP | Mouse | MGI:MGI:5584055|PMID:23603510 | |
| Biological Process | GO:0060976 | coronary vasculature development | Aplnr | IMP | Mouse | MGI:MGI:5914956|PMID:28890073 | |
| Biological Process | GO:0061371 | determination of heart left/right asymmetry | aplnra | IMP | ZFIN:ZDB-MRPHLNO-070525-6 | Zfish | PMID:31223282 |
| Biological Process | GO:0061371 | determination of heart left/right asymmetry | aplnrb | IMP | ZFIN:ZDB-MRPHLNO-070522-4 | Zfish | PMID:31223282 |
| Biological Process | GO:0007368 | determination of left/right symmetry | aplnra | IMP | ZFIN:ZDB-MRPHLNO-070525-6 | Zfish | PMID:31223282 |
| Biological Process | GO:0071910 | determination of liver left/right asymmetry | aplnra | IMP | ZFIN:ZDB-MRPHLNO-070525-6 | Zfish | PMID:31223282 |
| Biological Process | GO:0071910 | determination of liver left/right asymmetry | aplnrb | IMP | ZFIN:ZDB-MRPHLNO-070522-4 | Zfish | PMID:31223282 |
| Biological Process | GO:0003272 | endocardial cushion formation | Aplnr | IMP | Mouse | MGI:MGI:5584055|PMID:23603510 | |
| Biological Process | GO:0035987 | endodermal cell differentiation | aplnrb | IGI | ZFIN:ZDB-GENE-060929-512 | Zfish | PMID:27077952 |
| Biological Process | GO:0035987 | endodermal cell differentiation | aplnra | IGI | ZFIN:ZDB-GENE-050913-90 | Zfish | PMID:27077952 |
| Biological Process | GO:0055113 | epiboly involved in gastrulation with mouth forming second | aplnra | IMP | ZFIN:ZDB-MRPHLNO-110413-2 | Zfish | PMID:19922670 |
| Biological Process | GO:0055113 | epiboly involved in gastrulation with mouth forming second | aplnra | IMP | ZFIN:ZDB-MRPHLNO-110413-7 | Zfish | PMID:19922670 |
| Biological Process | GO:0055113 | epiboly involved in gastrulation with mouth forming second | aplnra | IMP | ZFIN:ZDB-MRPHLNO-110413-3 | Zfish | PMID:19922670 |
| Biological Process | GO:0090162 | establishment of epithelial cell polarity | aplnrb | IMP | ZFIN:ZDB-GENO-170413-7 | Zfish | PMID:27248505 |
| Biological Process | GO:0007186 | G protein-coupled receptor signaling pathway | APLNR | IMP | Human | PMID:25639753 | |
| Biological Process | GO:0007186 | G protein-coupled receptor signaling pathway | Aplnr | IMP | Rat | PMID:11359874|RGD:12907555 | |
| Biological Process | GO:0007369 | gastrulation | aplnrb | IMP | Zfish | PMID:17336905 | |
| Biological Process | GO:0001702 | gastrulation with mouth forming second | aplnrb | IMP | Zfish | PMID:17336905|ZFIN:ZDB-PUB-070310-8 | |
| Biological Process | GO:0007507 | heart development | aplnra | IGI | ZFIN:ZDB-GENE-050913-90 | Zfish | PMID:17336906|ZFIN:ZDB-PUB-070310-9 |
| Biological Process | GO:0007507 | heart development | aplnrb | IMP | Zfish | PMID:17336905|ZFIN:ZDB-PUB-070310-8 | |
| Biological Process | GO:0007507 | heart development | aplnrb | IMP | Zfish | PMID:24316148|ZFIN:ZDB-PUB-140106-3 | |
| Biological Process | GO:0007507 | heart development | aplnrb | IMP | Zfish | PMID:17336906|ZFIN:ZDB-PUB-070310-9 | |
| Biological Process | GO:0007507 | heart development | aplnrb | IMP | ZFIN:ZDB-MRPHLNO-070522-4 | Zfish | PMID:17336905 |
| Biological Process | GO:0007507 | heart development | aplnrb | IMP | ZFIN:ZDB-GENO-070525-1 | Zfish | PMID:17336906 |
| Biological Process | GO:0007507 | heart development | aplnrb | IMP | ZFIN:ZDB-MRPHLNO-070531-1 | Zfish | PMID:17336906 |
| Biological Process | GO:0007507 | heart development | aplnrb | IGI | ZFIN:ZDB-GENE-060929-512 | Zfish | PMID:27077952 |
| Biological Process | GO:0007507 | heart development | aplnra | IGI | ZFIN:ZDB-GENE-050913-90 | Zfish | PMID:27077952 |
| Biological Process | GO:0001947 | heart looping | Aplnr | IMP | Mouse | MGI:MGI:5584055|PMID:23603510 | |
| Biological Process | GO:0070121 | Kupffer's vesicle development | aplnra | IMP | ZFIN:ZDB-MRPHLNO-070525-6 | Zfish | PMID:31223282 |
| Biological Process | GO:0001945 | lymph vessel development | aplnra | IMP | ZFIN:ZDB-MRPHLNO-140317-4 | Zfish | PMID:24311379 |
| Biological Process | GO:0008078 | mesodermal cell migration | aplnrb | IMP | ZFIN:ZDB-MRPHLNO-070531-1 | Zfish | PMID:17336906 |
| Biological Process | GO:0043951 | negative regulation of cAMP-mediated signaling | APLNR | IMP | Human | PMID:25639753 | |
| Biological Process | GO:0043951 | negative regulation of cAMP-mediated signaling | Aplnr | IMP | Rat | PMID:11359874|RGD:12907555 | |
| Biological Process | GO:0010629 | negative regulation of gene expression | Aplnr | IMP | Mouse | MGI:MGI:5584055|PMID:23603510 | |
| Biological Process | GO:0045766 | positive regulation of angiogenesis | APLNR | IMP | Human | PMID:25639753 | |
| Biological Process | GO:1903589 | positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis | Aplnr | IMP | Mouse | MGI:MGI:5914956|PMID:28890073 | |
| Biological Process | GO:0031065 | positive regulation of histone deacetylation | Aplnr | IMP | Mouse | MGI:MGI:5584055|PMID:23603510 | |
| Biological Process | GO:1904325 | positive regulation of inhibitory G protein-coupled receptor phosphorylation | APLNR | IMP | Human | PMID:25639753 | |
| Biological Process | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol | APLNR | IMP | Human | PMID:25639753 | |
| Biological Process | GO:0050878 | regulation of body fluid levels | Aplnr | IMP | Rat | PMID:12787050|RGD:1304389 | |
| Biological Process | GO:1903596 | regulation of gap junction assembly | Aplnr | IMP | Mouse | MGI:MGI:5584055|PMID:23603510 | |
| Biological Process | GO:0010468 | regulation of gene expression | Aplnr | IMP | Mouse | MGI:MGI:5584055|PMID:23603510 | |
| Biological Process | GO:1900107 | regulation of nodal signaling pathway | aplnrb | IGI | ZFIN:ZDB-GENE-060929-512 | Zfish | PMID:27077952 |
| Biological Process | GO:1900107 | regulation of nodal signaling pathway | aplnra | IGI | ZFIN:ZDB-GENE-050913-90 | Zfish | PMID:27077952 |
| Biological Process | GO:0002040 | sprouting angiogenesis | aplnra | IGI | ZFIN:ZDB-GENO-220906-5 | Zfish | PMID:32955436 |
| Biological Process | GO:0002040 | sprouting angiogenesis | aplnrb | IGI | ZFIN:ZDB-GENO-220906-5 | Zfish | PMID:32955436 |
| Biological Process | GO:0035886 | vascular associated smooth muscle cell differentiation | Aplnr | IMP | Mouse | MGI:MGI:5584055|PMID:23603510 | |
| Biological Process | GO:0001944 | vasculature development | Aplnr | IMP | Mouse | MGI:MGI:5584055|PMID:23603510 | |
| Biological Process | GO:0001570 | vasculogenesis | Aplnr | IMP | Mouse | MGI:MGI:5913613|PMID:28663440 | |
| Biological Process | GO:0060841 | venous blood vessel development | Aplnr | IMP | Mouse | MGI:MGI:5584055|PMID:23603510 | |
| Biological Process | GO:0060412 | ventricular septum morphogenesis | Aplnr | IMP | Mouse | MGI:MGI:5584055|PMID:23603510 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 06/12/2024 MGI 6.13 |

|
|
|
||