|
Source |
Alliance of Genome Resources |
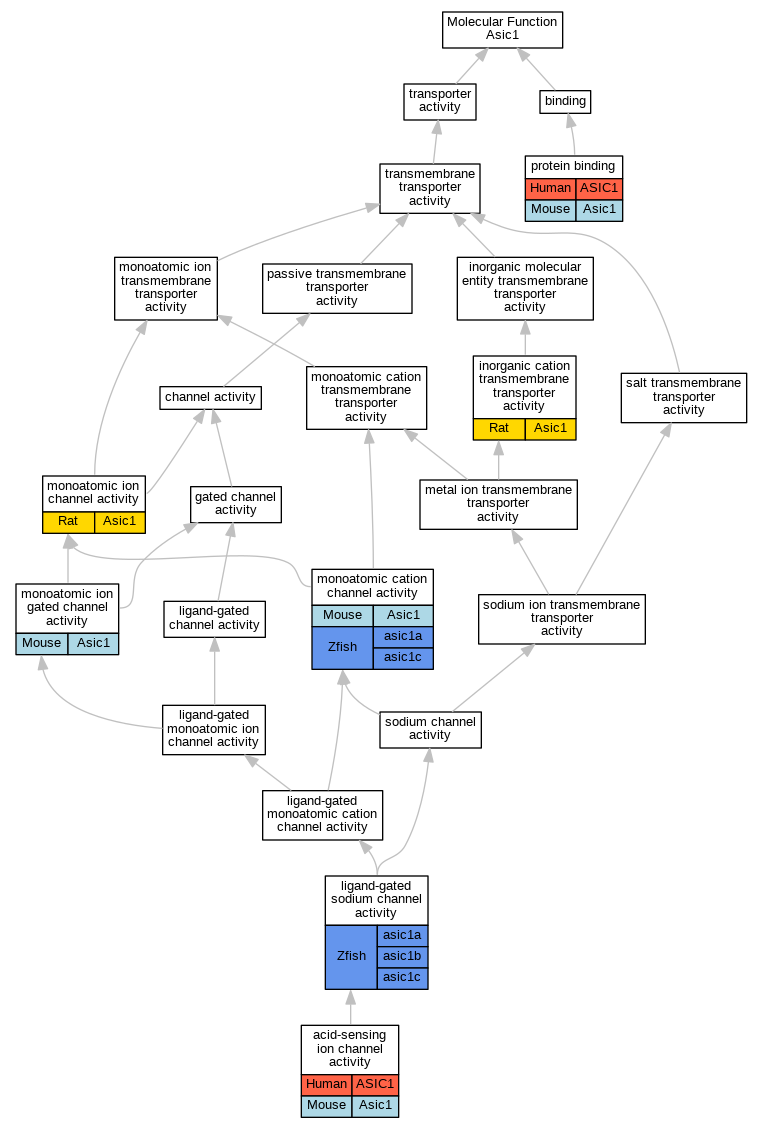
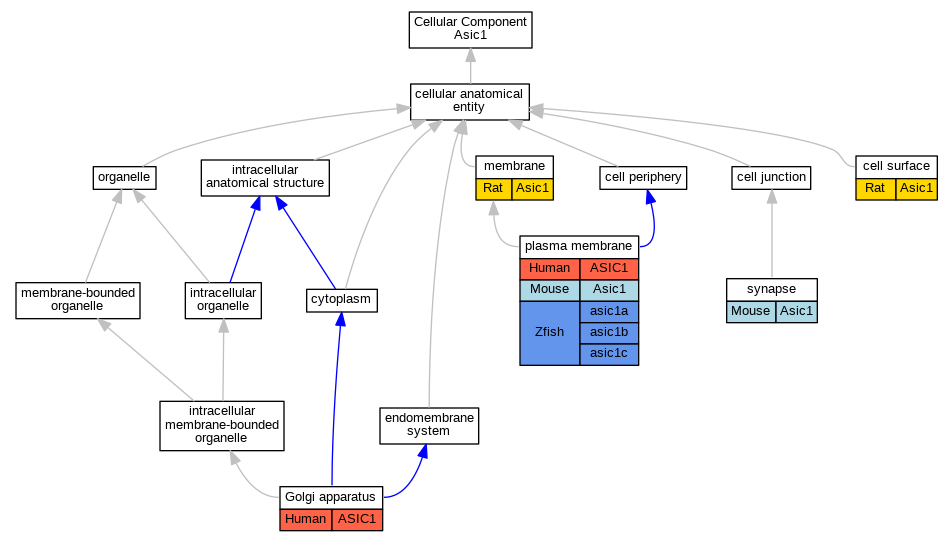
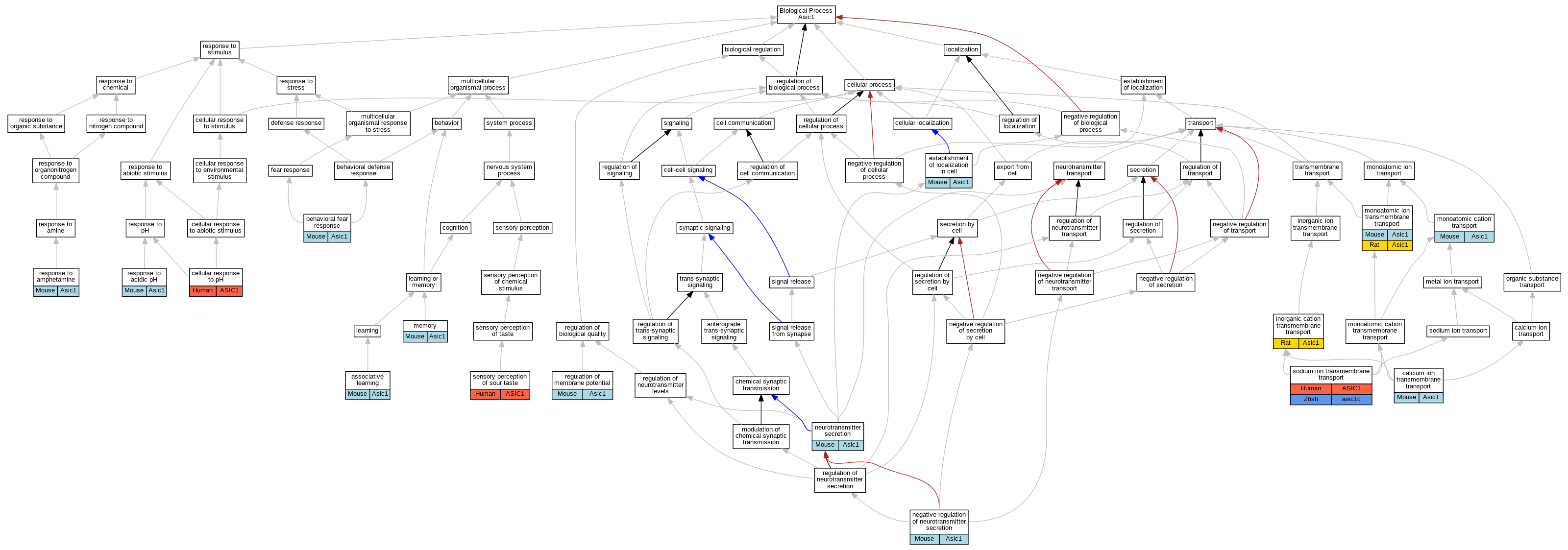
| Category | ID | Classification term | Gene | Evidence | Inferred from | Organism | Reference |
|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0044736 | acid-sensing ion channel activity | ASIC1 | IDA | Human | PMID:22760635 | |
| Molecular Function | GO:0044736 | acid-sensing ion channel activity | Asic1 | IDA | Mouse | PMID:24247984 | |
| Molecular Function | GO:0022890 | inorganic cation transmembrane transporter activity | Asic1 | IDA | Rat | PMID:11448963|RGD:70652 | |
| Molecular Function | GO:0015280 | ligand-gated sodium channel activity | asic1b | IDA | Zfish | PMID:14970195 | |
| Molecular Function | GO:0015280 | ligand-gated sodium channel activity | asic1a | IDA | Zfish | PMID:14970195 | |
| Molecular Function | GO:0015280 | ligand-gated sodium channel activity | asic1c | IDA | Zfish | PMID:14970195 | |
| Molecular Function | GO:0005261 | monoatomic cation channel activity | Asic1 | IDA | Mouse | PMID:11854527 | |
| Molecular Function | GO:0005261 | monoatomic cation channel activity | Asic1 | IMP | MGI:MGI:2179834 | Mouse | PMID:11854527 |
| Molecular Function | GO:0005261 | monoatomic cation channel activity | asic1a | IGI | ZFIN:ZDB-GENE-040513-3 | Zfish | PMID:14970195 |
| Molecular Function | GO:0005261 | monoatomic cation channel activity | asic1c | IPI | ZFIN:ZDB-GENE-040513-6 | Zfish | PMID:17686779 |
| Molecular Function | GO:0005261 | monoatomic cation channel activity | asic1c | IPI | ZFIN:ZDB-GENE-040513-5 | Zfish | PMID:17686779 |
| Molecular Function | GO:0005261 | monoatomic cation channel activity | asic1c | IGI | ZFIN:ZDB-GENE-040513-2 | Zfish | PMID:14970195 |
| Molecular Function | GO:0005216 | monoatomic ion channel activity | Asic1 | IDA | Rat | PMID:9707631|RGD:70653 | |
| Molecular Function | GO:0022839 | monoatomic ion gated channel activity | Asic1 | IMP | MGI:MGI:2179834 | Mouse | PMID:14960591 |
| Molecular Function | GO:0022839 | monoatomic ion gated channel activity | Asic1 | IDA | Mouse | PMID:14960591 | |
| Molecular Function | GO:0005515 | protein binding | ASIC1 | IPI | UniProtKB:Q9NRD5 | Human | PMID:11802773 |
| Molecular Function | GO:0005515 | protein binding | ASIC1 | IPI | UniProtKB:Q8N743 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | ASIC1 | IPI | UniProtKB:Q99735 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | Asic1 | IPI | UniProtKB:Q12791 | Mouse | MGI:MGI:6092628|PMID:18287010 |
| Molecular Function | GO:0005515 | protein binding | Asic1 | IPI | PR:Q925H0 | Mouse | PMID:19571134 |
| Cellular Component | GO:0009986 | cell surface | Asic1 | IDA | Rat | PMID:19074149|RGD:7242197 | |
| Cellular Component | GO:0005794 | Golgi apparatus | ASIC1 | IDA | Human | GO_REF:0000052 | |
| Cellular Component | GO:0016020 | membrane | Asic1 | IDA | Rat | PMID:11448963|RGD:70652 | |
| Cellular Component | GO:0005886 | plasma membrane | ASIC1 | IDA | Human | GO_REF:0000052 | |
| Cellular Component | GO:0005886 | plasma membrane | ASIC1 | IMP | Human | PMID:22760635 | |
| Cellular Component | GO:0005886 | plasma membrane | Asic1 | IDA | Mouse | PMID:11854527 | |
| Cellular Component | GO:0005886 | plasma membrane | asic1b | IDA | Zfish | PMID:14970195 | |
| Cellular Component | GO:0005886 | plasma membrane | asic1a | IDA | Zfish | PMID:14970195 | |
| Cellular Component | GO:0005886 | plasma membrane | asic1c | IPI | ZFIN:ZDB-GENE-040513-6 | Zfish | PMID:17686779 |
| Cellular Component | GO:0005886 | plasma membrane | asic1c | IDA | Zfish | PMID:14970195 | |
| Cellular Component | GO:0005886 | plasma membrane | asic1c | IPI | ZFIN:ZDB-GENE-040513-5 | Zfish | PMID:17686779 |
| Cellular Component | GO:0045202 | synapse | Asic1 | IDA | Mouse | PMID:11988176 | |
| Biological Process | GO:0008306 | associative learning | Asic1 | IMP | MGI:MGI:2179834 | Mouse | PMID:11988176 |
| Biological Process | GO:0001662 | behavioral fear response | Asic1 | IMP | Mouse | PMID:25828470 | |
| Biological Process | GO:0070588 | calcium ion transmembrane transport | Asic1 | IMP | MGI:MGI:2179834 | Mouse | PMID:19376200 |
| Biological Process | GO:0071467 | cellular response to pH | ASIC1 | IDA | Human | PMID:22760635 | |
| Biological Process | GO:0051649 | establishment of localization in cell | Asic1 | IMP | MGI:MGI:2179834 | Mouse | PMID:15470133 |
| Biological Process | GO:0051649 | establishment of localization in cell | Asic1 | IMP | MGI:MGI:2179834 | Mouse | PMID:14960591 |
| Biological Process | GO:0098662 | inorganic cation transmembrane transport | Asic1 | IDA | Rat | PMID:11448963|RGD:70652 | |
| Biological Process | GO:0007613 | memory | Asic1 | IMP | MGI:MGI:2179834 | Mouse | PMID:11988176 |
| Biological Process | GO:0006812 | monoatomic cation transport | Asic1 | IMP | MGI:MGI:2179834 | Mouse | PMID:11854527 |
| Biological Process | GO:0006812 | monoatomic cation transport | Asic1 | IDA | Mouse | PMID:11854527 | |
| Biological Process | GO:0034220 | monoatomic ion transmembrane transport | Asic1 | IMP | MGI:MGI:2179834 | Mouse | PMID:19376200 |
| Biological Process | GO:0034220 | monoatomic ion transmembrane transport | Asic1 | IMP | MGI:MGI:2179834 | Mouse | PMID:14960591 |
| Biological Process | GO:0034220 | monoatomic ion transmembrane transport | Asic1 | IDA | Mouse | PMID:14960591 | |
| Biological Process | GO:0034220 | monoatomic ion transmembrane transport | Asic1 | IMP | MGI:MGI:2179834 | Mouse | PMID:15470133 |
| Biological Process | GO:0034220 | monoatomic ion transmembrane transport | Asic1 | IMP | Rat | PMID:19074149|RGD:7242197 | |
| Biological Process | GO:0046929 | negative regulation of neurotransmitter secretion | Asic1 | IMP | MGI:MGI:2179834 | Mouse | PMID:18094106 |
| Biological Process | GO:0007269 | neurotransmitter secretion | Asic1 | IMP | MGI:MGI:2179834 | Mouse | PMID:18094106 |
| Biological Process | GO:0042391 | regulation of membrane potential | Asic1 | IMP | MGI:MGI:2179834 | Mouse | PMID:19376200 |
| Biological Process | GO:0010447 | response to acidic pH | Asic1 | IMP | MGI:MGI:2179834 | Mouse | PMID:11854527 |
| Biological Process | GO:0010447 | response to acidic pH | Asic1 | IMP | MGI:MGI:2179834 | Mouse | PMID:11988176 |
| Biological Process | GO:0010447 | response to acidic pH | Asic1 | IDA | Mouse | PMID:11854527 | |
| Biological Process | GO:0001975 | response to amphetamine | Asic1 | IMP | Mouse | PMID:25828470 | |
| Biological Process | GO:0050915 | sensory perception of sour taste | ASIC1 | IMP | Human | PMID:19812697 | |
| Biological Process | GO:0035725 | sodium ion transmembrane transport | ASIC1 | IDA | Human | PMID:22760635 | |
| Biological Process | GO:0035725 | sodium ion transmembrane transport | asic1c | IDA | Zfish | PMID:14970195|ZFIN:ZDB-PUB-040225-4 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 07/05/2024 MGI 6.24 |

|
|
|
||