|
Source |
Alliance of Genome Resources |
| Category | ID | Classification term | Gene | Evidence | Inferred from | Organism | Reference |
|---|---|---|---|---|---|---|---|
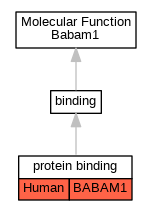
| Molecular Function | GO:0005515 | protein binding | BABAM1 | IPI | UniProtKB:Q9NXR7 | Human | PMID:16189514 |
| Molecular Function | GO:0005515 | protein binding | BABAM1 | IPI | UniProtKB:Q9NXR7 | Human | PMID:19261748 |
| Molecular Function | GO:0005515 | protein binding | BABAM1 | IPI | UniProtKB:Q6UWZ7 | Human | PMID:19261749 |
| Molecular Function | GO:0005515 | protein binding | BABAM1 | IPI | UniProtKB:Q9NXR7 | Human | PMID:19615732 |
| Molecular Function | GO:0005515 | protein binding | BABAM1 | IPI | UniProtKB:Q9NYB0 | Human | PMID:21044950 |
| Molecular Function | GO:0005515 | protein binding | BABAM1 | IPI | UniProtKB:Q9H2K2 | Human | PMID:22153077 |
| Molecular Function | GO:0005515 | protein binding | BABAM1 | IPI | UniProtKB:P14373 | Human | PMID:25416956 |
| Molecular Function | GO:0005515 | protein binding | BABAM1 | IPI | UniProtKB:Q9H2K2 | Human | PMID:28514442 |
| Molecular Function | GO:0005515 | protein binding | BABAM1 | IPI | UniProtKB:Q9NXR7 | Human | PMID:28514442 |
| Molecular Function | GO:0005515 | protein binding | BABAM1 | IPI | UniProtKB:P14373 | Human | PMID:31515488 |
| Molecular Function | GO:0005515 | protein binding | BABAM1 | IPI | UniProtKB:Q9NXR7 | Human | PMID:31515488 |
| Molecular Function | GO:0005515 | protein binding | BABAM1 | IPI | UniProtKB:P14373 | Human | PMID:32296183 |
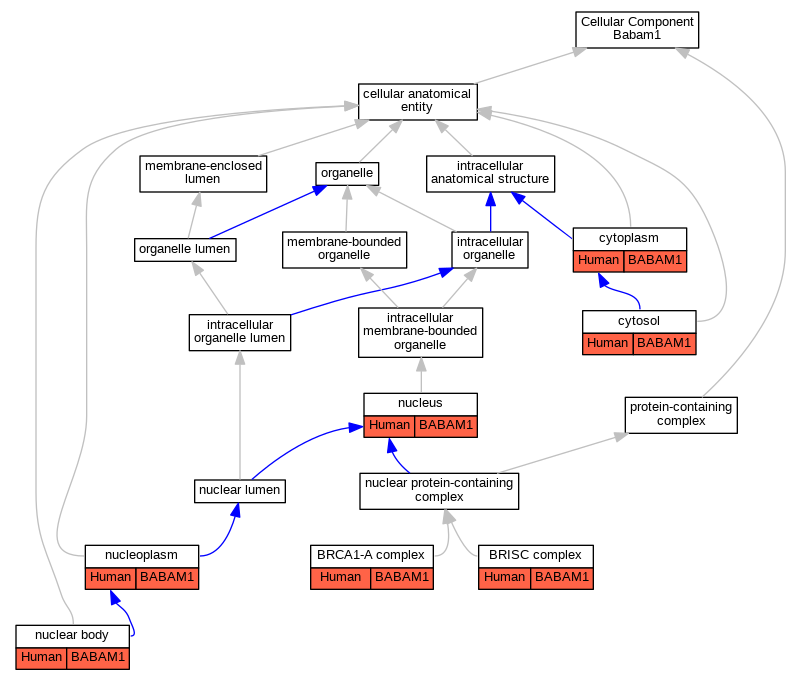
| Cellular Component | GO:0070531 | BRCA1-A complex | BABAM1 | IDA | Human | PMID:19261746 | |
| Cellular Component | GO:0070531 | BRCA1-A complex | BABAM1 | IDA | Human | PMID:19261748 | |
| Cellular Component | GO:0070531 | BRCA1-A complex | BABAM1 | IDA | Human | PMID:19261749 | |
| Cellular Component | GO:0070552 | BRISC complex | BABAM1 | IDA | Human | PMID:19214193 | |
| Cellular Component | GO:0070552 | BRISC complex | BABAM1 | IDA | Human | PMID:24075985 | |
| Cellular Component | GO:0005737 | cytoplasm | BABAM1 | IDA | Human | PMID:19261749 | |
| Cellular Component | GO:0005737 | cytoplasm | BABAM1 | IDA | Human | PMID:24075985 | |
| Cellular Component | GO:0005829 | cytosol | BABAM1 | IDA | Human | GO_REF:0000052 | |
| Cellular Component | GO:0016604 | nuclear body | BABAM1 | IDA | Human | GO_REF:0000052 | |
| Cellular Component | GO:0005654 | nucleoplasm | BABAM1 | IDA | Human | GO_REF:0000052 | |
| Cellular Component | GO:0005634 | nucleus | BABAM1 | IDA | Human | PMID:19261746 | |
| Cellular Component | GO:0005634 | nucleus | BABAM1 | IDA | Human | PMID:19261748 | |
| Cellular Component | GO:0005634 | nucleus | BABAM1 | IDA | Human | PMID:19261749 | |
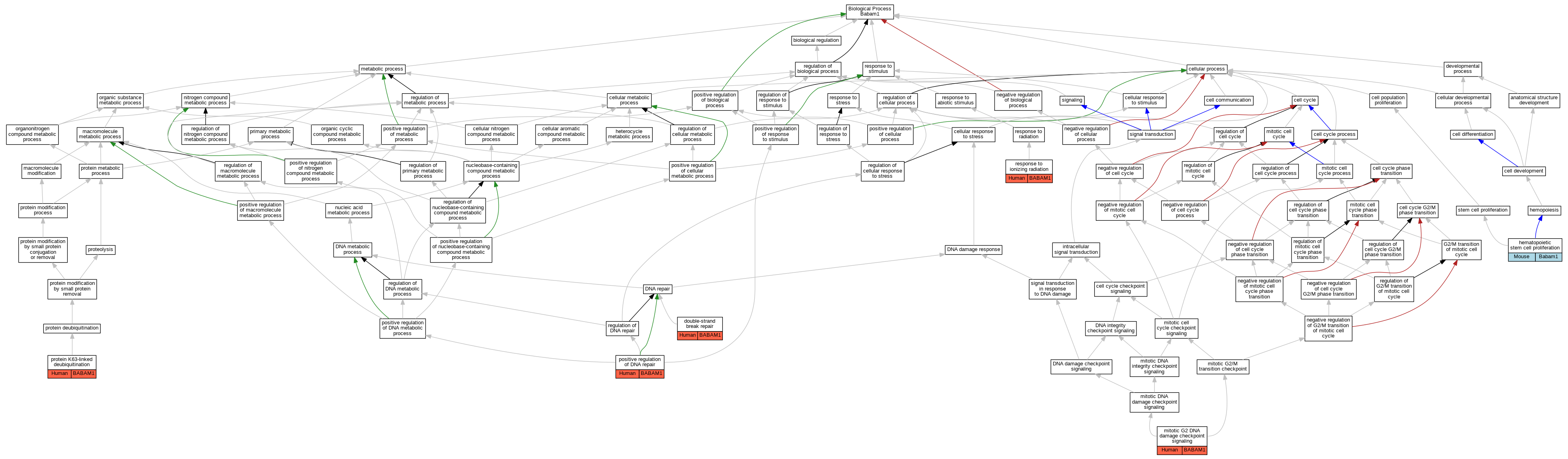
| Biological Process | GO:0006302 | double-strand break repair | BABAM1 | IMP | Human | PMID:19261746 | |
| Biological Process | GO:0006302 | double-strand break repair | BABAM1 | IMP | Human | PMID:19261748 | |
| Biological Process | GO:0006302 | double-strand break repair | BABAM1 | IMP | Human | PMID:19261749 | |
| Biological Process | GO:0071425 | hematopoietic stem cell proliferation | Babam1 | IMP | MGI:MGI:3994220 | Mouse | PMID:25636339 |
| Biological Process | GO:0007095 | mitotic G2 DNA damage checkpoint signaling | BABAM1 | IMP | Human | PMID:19261746 | |
| Biological Process | GO:0007095 | mitotic G2 DNA damage checkpoint signaling | BABAM1 | IMP | Human | PMID:19261748 | |
| Biological Process | GO:0007095 | mitotic G2 DNA damage checkpoint signaling | BABAM1 | IMP | Human | PMID:19261749 | |
| Biological Process | GO:0045739 | positive regulation of DNA repair | BABAM1 | IMP | Human | PMID:19261746 | |
| Biological Process | GO:0045739 | positive regulation of DNA repair | BABAM1 | IMP | Human | PMID:19261748 | |
| Biological Process | GO:0045739 | positive regulation of DNA repair | BABAM1 | IMP | Human | PMID:19261749 | |
| Biological Process | GO:0070536 | protein K63-linked deubiquitination | BABAM1 | IMP | Human | PMID:19261746 | |
| Biological Process | GO:0010212 | response to ionizing radiation | BABAM1 | IMP | Human | PMID:19261748 | |
| Biological Process | GO:0010212 | response to ionizing radiation | BABAM1 | IMP | Human | PMID:19261749 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 09/03/2024 MGI 6.24 |

|
|
|
||