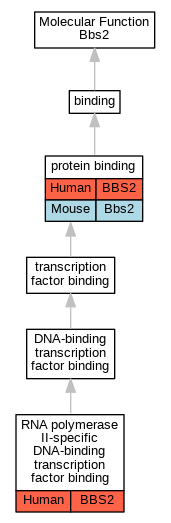
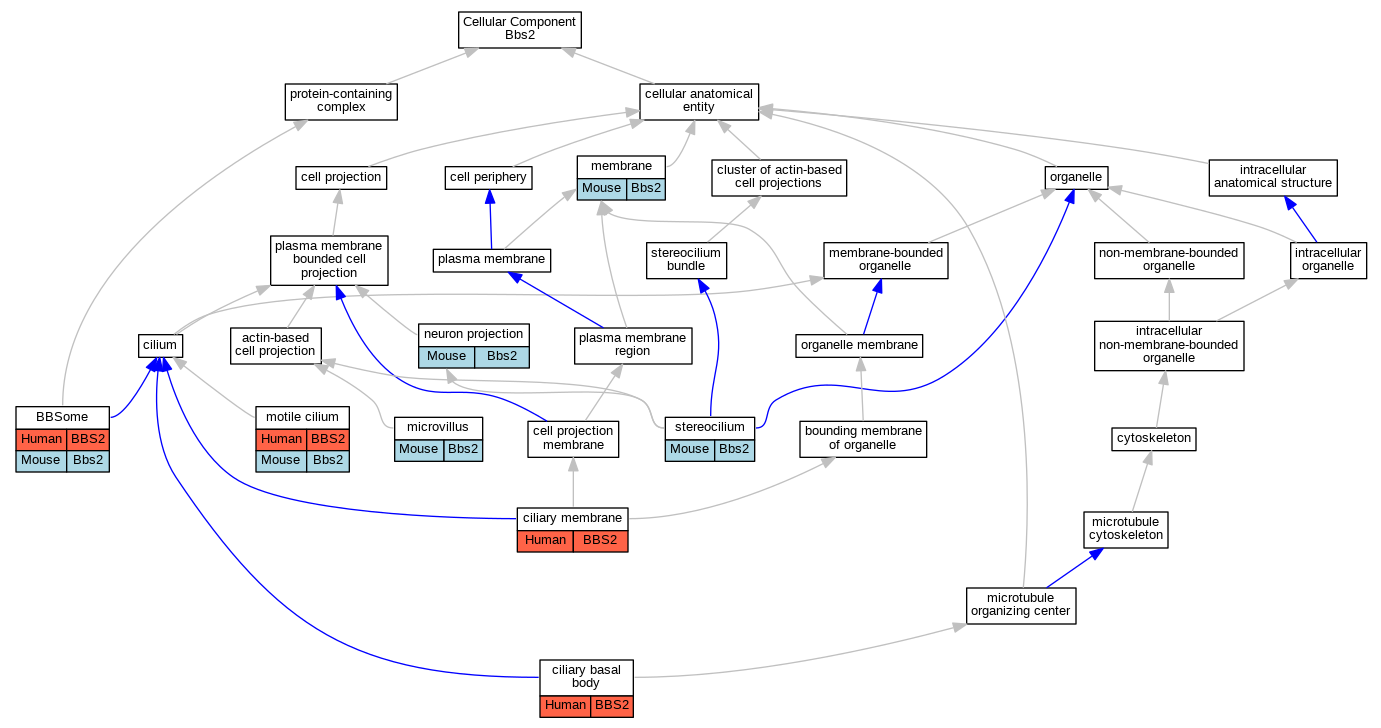
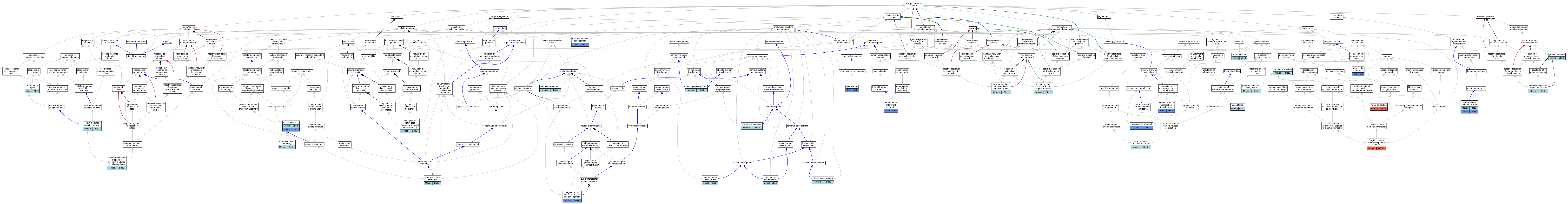
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0005515 | protein binding | BBS2 | IPI | UniProtKB:P61289 | Human | PMID:16189514 |
| Molecular Function | GO:0005515 | protein binding | BBS2 | IPI | UniProtKB:Q9BUN5 | Human | PMID:16327777 |
| Molecular Function | GO:0005515 | protein binding | BBS2 | IPI | UniProtKB:Q3SYG4 | Human | PMID:17574030 |
| Molecular Function | GO:0005515 | protein binding | BBS2 | IPI | UniProtKB:Q8IWZ6 | Human | PMID:17574030 |
| Molecular Function | GO:0005515 | protein binding | BBS2 | IPI | UniProtKB:Q8NFJ9 | Human | PMID:17574030 |
| Molecular Function | GO:0005515 | protein binding | BBS2 | IPI | UniProtKB:P05062 | Human | PMID:18000879 |
| Molecular Function | GO:0005515 | protein binding | BBS2 | IPI | UniProtKB:P68104 | Human | PMID:18000879 |
| Molecular Function | GO:0005515 | protein binding | BBS2 | IPI | UniProtKB:Q15154 | Human | PMID:18000879 |
| Molecular Function | GO:0005515 | protein binding | BBS2 | IPI | UniProtKB:Q9NRI5 | Human | PMID:18762586 |
| Molecular Function | GO:0005515 | protein binding | BBS2 | IPI | UniProtKB:Q3SYG4 | Human | PMID:20080638 |
| Molecular Function | GO:0005515 | protein binding | BBS2 | IPI | UniProtKB:Q8IWZ6 | Human | PMID:20080638 |
| Molecular Function | GO:0005515 | protein binding | BBS2 | IPI | UniProtKB:Q9NPJ1 | Human | PMID:20080638 |
| Molecular Function | GO:0005515 | protein binding | BBS2 | IPI | UniProtKB:Q3SYG4 | Human | PMID:22139371 |
| Molecular Function | GO:0005515 | protein binding | BBS2 | IPI | UniProtKB:Q3SYG4 | Human | PMID:22500027 |
| Molecular Function | GO:0005515 | protein binding | BBS2 | IPI | UniProtKB:Q8IWZ6 | Human | PMID:22500027 |
| Molecular Function | GO:0005515 | protein binding | BBS2 | IPI | UniProtKB:Q8NFJ9 | Human | PMID:22500027 |
| Molecular Function | GO:0005515 | protein binding | BBS2 | IPI | UniProtKB:Q9NPJ1 | Human | PMID:22500027 |
| Molecular Function | GO:0005515 | protein binding | BBS2 | IPI | UniProtKB:P61289 | Human | PMID:25416956 |
| Molecular Function | GO:0005515 | protein binding | BBS2 | IPI | UniProtKB:Q93062 | Human | PMID:25416956 |
| Molecular Function | GO:0005515 | protein binding | BBS2 | IPI | UniProtKB:Q99750 | Human | PMID:25416956 |
| Molecular Function | GO:0005515 | protein binding | BBS2 | IPI | UniProtKB:Q15051 | Human | PMID:25552655 |
| Molecular Function | GO:0005515 | protein binding | BBS2 | IPI | UniProtKB:Q3SYG4 | Human | PMID:27173435 |
| Molecular Function | GO:0005515 | protein binding | BBS2 | IPI | UniProtKB:Q8IWZ6 | Human | PMID:27173435 |
| Molecular Function | GO:0005515 | protein binding | BBS2 | IPI | UniProtKB:Q8NFJ9 | Human | PMID:27173435 |
| Molecular Function | GO:0005515 | protein binding | BBS2 | IPI | UniProtKB:Q8IWZ6 | Human | PMID:28514442 |
| Molecular Function | GO:0005515 | protein binding | BBS2 | IPI | UniProtKB:Q3SYG4 | Human | PMID:29039417 |
| Molecular Function | GO:0005515 | protein binding | BBS2 | IPI | UniProtKB:Q8IWZ6-2 | Human | PMID:29039417 |
| Molecular Function | GO:0005515 | protein binding | BBS2 | IPI | UniProtKB:P52597 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | BBS2 | IPI | UniProtKB:P61289 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | BBS2 | IPI | UniProtKB:P61968 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | BBS2 | IPI | UniProtKB:Q16656-4 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | BBS2 | IPI | UniProtKB:Q53EP0-3 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | BBS2 | IPI | UniProtKB:Q99750 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | BBS2 | IPI | UniProtKB:Q9Y238 | Human | PMID:33144677 |
| Molecular Function | GO:0005515 | protein binding | Bbs2 | IPI | PR:Q8BMD2 | Mouse | PMID:27979967 |
| Molecular Function | GO:0061629 | RNA polymerase II-specific DNA-binding transcription factor binding | BBS2 | IPI | UniProtKB:Q99496 | Human | PMID:22302990 |
| Cellular Component | GO:0034464 | BBSome | BBS2 | IPI | | Human | PMID:19081074 |
| Cellular Component | GO:0034464 | BBSome | BBS2 | IDA | | Human | PMID:17574030 |
| Cellular Component | GO:0034464 | BBSome | BBS2 | IDA | | Human | PMID:24550735 |
| Cellular Component | GO:0034464 | BBSome | Bbs2 | IDA | | Mouse | MGI:MGI:5560383|PMID:24550735 |
| Cellular Component | GO:0034464 | BBSome | Bbs2 | IDA | | Mouse | PMID:20080638 |
| Cellular Component | GO:0034464 | BBSome | Bbs2 | IDA | | Mouse | PMID:22500027 |
| Cellular Component | GO:0034464 | BBSome | Bbs2 | IDA | | Mouse | PMID:22072986 |
| Cellular Component | GO:0036064 | ciliary basal body | BBS2 | IDA | | Human | PMID:18299575 |
| Cellular Component | GO:0060170 | ciliary membrane | BBS2 | IDA | | Human | PMID:19081074 |
| Cellular Component | GO:0016020 | membrane | Bbs2 | IDA | | Mouse | PMID:22139371 |
| Cellular Component | GO:0005902 | microvillus | Bbs2 | IDA | | Mouse | PMID:25605782 |
| Cellular Component | GO:0031514 | motile cilium | BBS2 | IDA | | Human | PMID:18299575 |
| Cellular Component | GO:0031514 | motile cilium | Bbs2 | IMP | | Mouse | MGI:MGI:3776933|PMID:18299575 |
| Cellular Component | GO:0043005 | neuron projection | Bbs2 | IDA | | Mouse | PMID:25605782 |
| Cellular Component | GO:0032420 | stereocilium | Bbs2 | IDA | | Mouse | PMID:25605782 |
| Biological Process | GO:0030534 | adult behavior | Bbs2 | IMP | MGI:MGI:3521669 | Mouse | PMID:15539463 |
| Biological Process | GO:0014824 | artery smooth muscle contraction | Bbs2 | IMP | MGI:MGI:3521669 | Mouse | PMID:20852044 |
| Biological Process | GO:0048854 | brain morphogenesis | Bbs2 | IMP | MGI:MGI:3521669 | Mouse | PMID:18032602 |
| Biological Process | GO:0051216 | cartilage development | Bbs2 | IMP | MGI:MGI:3521669 | Mouse | PMID:19195025 |
| Biological Process | GO:0021987 | cerebral cortex development | Bbs2 | IMP | MGI:MGI:3521669 | Mouse | PMID:18032602 |
| Biological Process | GO:0060271 | cilium assembly | Bbs2 | IMP | | Mouse | MGI:MGI:3776933|PMID:18299575 |
| Biological Process | GO:0060271 | cilium assembly | bbs2 | IMP | | Zfish | PMID:16399798|ZFIN:ZDB-PUB-060124-4 |
| Biological Process | GO:0060271 | cilium assembly | bbs2 | IMP | ZFIN:ZDB-MRPHLNO-060209-3 | Zfish | PMID:16399798 |
| Biological Process | GO:0007368 | determination of left/right symmetry | bbs2 | IGI | ZFIN:ZDB-MRPHLNO-060209-3,ZFIN:ZDB-MRPHLNO-081027-1 | Zfish | PMID:18381349 |
| Biological Process | GO:0007368 | determination of left/right symmetry | bbs2 | IGI | ZFIN:ZDB-MRPHLNO-060209-3,ZFIN:ZDB-MRPHLNO-081027-3 | Zfish | PMID:18381349 |
| Biological Process | GO:0007368 | determination of left/right symmetry | bbs2 | IGI | ZFIN:ZDB-MRPHLNO-060209-3,ZFIN:ZDB-MRPHLNO-060519-1 | Zfish | PMID:18381349 |
| Biological Process | GO:0007368 | determination of left/right symmetry | bbs2 | IGI | ZFIN:ZDB-MRPHLNO-060209-3,ZFIN:ZDB-MRPHLNO-060209-5 | Zfish | PMID:18381349 |
| Biological Process | GO:0045444 | fat cell differentiation | Bbs2 | IEP | | Mouse | MGI:MGI:3794016|PMID:17379567 |
| Biological Process | GO:0007369 | gastrulation | bbs2 | IMP | ZFIN:ZDB-MRPHLNO-131205-1 | Zfish | PMID:20498079 |
| Biological Process | GO:0010467 | gene expression | Bbs2 | IMP | MGI:MGI:3521669 | Mouse | PMID:19150989 |
| Biological Process | GO:0010467 | gene expression | Bbs2 | IMP | MGI:MGI:3521669 | Mouse | PMID:18317593 |
| Biological Process | GO:0043001 | Golgi to plasma membrane protein transport | BBS2 | IMP | | Human | PMID:19150989 |
| Biological Process | GO:0021766 | hippocampus development | Bbs2 | IMP | MGI:MGI:3521669 | Mouse | PMID:18032602 |
| Biological Process | GO:0046907 | intracellular transport | bbs2 | IMP | ZFIN:ZDB-MRPHLNO-060209-3 | Zfish | PMID:16399798 |
| Biological Process | GO:0070121 | Kupffer's vesicle development | bbs2 | IGI | ZFIN:ZDB-MRPHLNO-060209-3,ZFIN:ZDB-MRPHLNO-060209-5 | Zfish | PMID:18381349 |
| Biological Process | GO:0070121 | Kupffer's vesicle development | bbs2 | IGI | ZFIN:ZDB-MRPHLNO-060209-3,ZFIN:ZDB-MRPHLNO-081027-3 | Zfish | PMID:18381349 |
| Biological Process | GO:0070121 | Kupffer's vesicle development | bbs2 | IGI | ZFIN:ZDB-MRPHLNO-060209-3,ZFIN:ZDB-MRPHLNO-081027-1 | Zfish | PMID:18381349 |
| Biological Process | GO:0070121 | Kupffer's vesicle development | bbs2 | IMP | ZFIN:ZDB-MRPHLNO-060209-3 | Zfish | PMID:16399798 |
| Biological Process | GO:0070121 | Kupffer's vesicle development | bbs2 | IGI | ZFIN:ZDB-MRPHLNO-060209-3,ZFIN:ZDB-MRPHLNO-060519-1 | Zfish | PMID:18381349 |
| Biological Process | GO:0033210 | leptin-mediated signaling pathway | Bbs2 | IMP | MGI:MGI:3521669 | Mouse | PMID:19150989 |
| Biological Process | GO:0032402 | melanosome transport | bbs2 | IMP | | Zfish | PMID:16399798|ZFIN:ZDB-PUB-060124-4 |
| Biological Process | GO:0032402 | melanosome transport | bbs2 | IGI | ZFIN:ZDB-MRPHLNO-060209-3,ZFIN:ZDB-MRPHLNO-060209-5 | Zfish | PMID:18381349 |
| Biological Process | GO:0032402 | melanosome transport | bbs2 | IGI | ZFIN:ZDB-MRPHLNO-060209-3,ZFIN:ZDB-MRPHLNO-081027-1 | Zfish | PMID:18381349 |
| Biological Process | GO:0032402 | melanosome transport | bbs2 | IGI | ZFIN:ZDB-MRPHLNO-060209-3,ZFIN:ZDB-MRPHLNO-081027-3 | Zfish | PMID:18381349 |
| Biological Process | GO:0032402 | melanosome transport | bbs2 | IGI | ZFIN:ZDB-MRPHLNO-060209-3,ZFIN:ZDB-MRPHLNO-060519-1 | Zfish | PMID:18381349 |
| Biological Process | GO:0032402 | melanosome transport | bbs2 | IMP | ZFIN:ZDB-MRPHLNO-060209-3 | Zfish | PMID:16399798 |
| Biological Process | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway | Bbs2 | IMP | | Mouse | MGI:MGI:3836843|PMID:19150989 |
| Biological Process | GO:0010629 | negative regulation of gene expression | Bbs2 | IMP | MGI:MGI:3521669 | Mouse | PMID:18317593 |
| Biological Process | GO:0010629 | negative regulation of gene expression | Bbs2 | IMP | MGI:MGI:3521669 | Mouse | PMID:19150989 |
| Biological Process | GO:0040015 | negative regulation of multicellular organism growth | Bbs2 | IMP | MGI:MGI:3521669 | Mouse | PMID:15539463 |
| Biological Process | GO:1905515 | non-motile cilium assembly | Bbs2 | IMP | MGI:MGI:3521669 | Mouse | PMID:19195025 |
| Biological Process | GO:0045494 | photoreceptor cell maintenance | Bbs2 | IMP | MGI:MGI:3521669 | Mouse | PMID:15539463 |
| Biological Process | GO:0045494 | photoreceptor cell maintenance | bbs2 | IMP | ZFIN:ZDB-GENO-210812-5 | Zfish | PMID:33324636 |
| Biological Process | GO:0051877 | pigment granule aggregation in cell center | bbs2 | IMP | ZFIN:ZDB-MRPHLNO-060209-3 | Zfish | PMID:16399798 |
| Biological Process | GO:0040018 | positive regulation of multicellular organism growth | Bbs2 | IMP | MGI:MGI:3521669 | Mouse | PMID:18317593 |
| Biological Process | GO:0008104 | protein localization | Bbs2 | IMP | MGI:MGI:3521669 | Mouse | PMID:15539463 |
| Biological Process | GO:0008104 | protein localization | Bbs2 | IMP | MGI:MGI:3521669 | Mouse | PMID:22228099 |
| Biological Process | GO:0033365 | protein localization to organelle | Bbs2 | IDA | | Mouse | MGI:MGI:3778303|PMID:18334641 |
| Biological Process | GO:0060296 | regulation of cilium beat frequency involved in ciliary motility | Bbs2 | IMP | | Mouse | MGI:MGI:3776933|PMID:18299575 |
| Biological Process | GO:0042478 | regulation of eye photoreceptor cell development | bbs2 | IMP | ZFIN:ZDB-GENO-210812-5 | Zfish | PMID:33324636 |
| Biological Process | GO:0044321 | response to leptin | Bbs2 | IMP | MGI:MGI:3521669 | Mouse | PMID:18317593 |
| Biological Process | GO:0007288 | sperm axoneme assembly | Bbs2 | IMP | MGI:MGI:3521669 | Mouse | PMID:15539463 |
| Biological Process | GO:0021756 | striatum development | Bbs2 | IMP | MGI:MGI:3521669 | Mouse | PMID:18032602 |
| Biological Process | GO:0042311 | vasodilation | Bbs2 | IMP | MGI:MGI:3521669 | Mouse | PMID:20852044 |
| Biological Process | GO:0007601 | visual perception | BBS2 | IMP | | Human | PMID:25541840 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
 Analysis Tools
Analysis Tools