| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
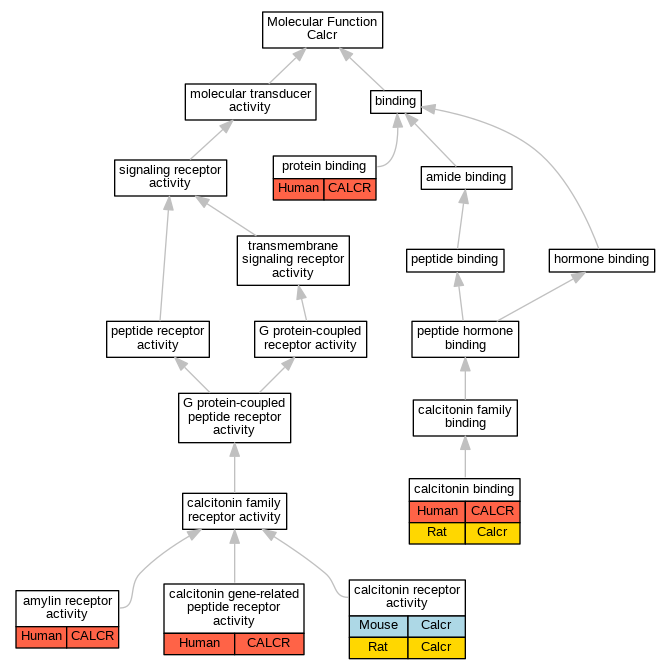
| Molecular Function | GO:0097643 | amylin receptor activity | CALCR | IGI | UniProtKB:O60894,UniProtKB:P06881|UniProtKB:O60894,UniProtKB:P10997:PRO_0000004106|UniProtKB:O60894,UniProtKB:P12969:PRO_0000004122|UniProtKB:O60894,UniProtKB:P35318|UniProtKB:O60895,UniProtKB:P12969:PRO_0000004122|UniProtKB:O60895,UniProtKB:P35318|UniProtKB:O60896,UniProtKB:P10997:PRO_0000004106 | Human | PMID:22500019 |
| Molecular Function | GO:0097643 | amylin receptor activity | CALCR | IPI | UniProtKB:O60894 | Human | PMID:14722252 |
| Molecular Function | GO:0032841 | calcitonin binding | CALCR | IPI | UniProtKB:P01263 | Human | PMID:15563840 |
| Molecular Function | GO:0032841 | calcitonin binding | CALCR | IDA | | Human | PMID:8078488 |
| Molecular Function | GO:0032841 | calcitonin binding | Calcr | IDA | | Rat | PMID:7723741|RGD:2303052 |
| Molecular Function | GO:0001635 | calcitonin gene-related peptide receptor activity | CALCR | IPI | UniProtKB:O60894 | Human | PMID:14722252 |
| Molecular Function | GO:0004948 | calcitonin receptor activity | Calcr | IDA | | Mouse | PMID:7988453 |
| Molecular Function | GO:0004948 | calcitonin receptor activity | Calcr | IDA | | Rat | PMID:7723741|RGD:2303052 |
| Molecular Function | GO:0004948 | calcitonin receptor activity | Calcr | IDA | | Rat | PMID:8395656|RGD:727711 |
| Molecular Function | GO:0005515 | protein binding | CALCR | IPI | UniProtKB:P06881 | Human | PMID:14722252 |
| Molecular Function | GO:0005515 | protein binding | CALCR | IPI | UniProtKB:Q91157 | Human | PMID:7588285 |
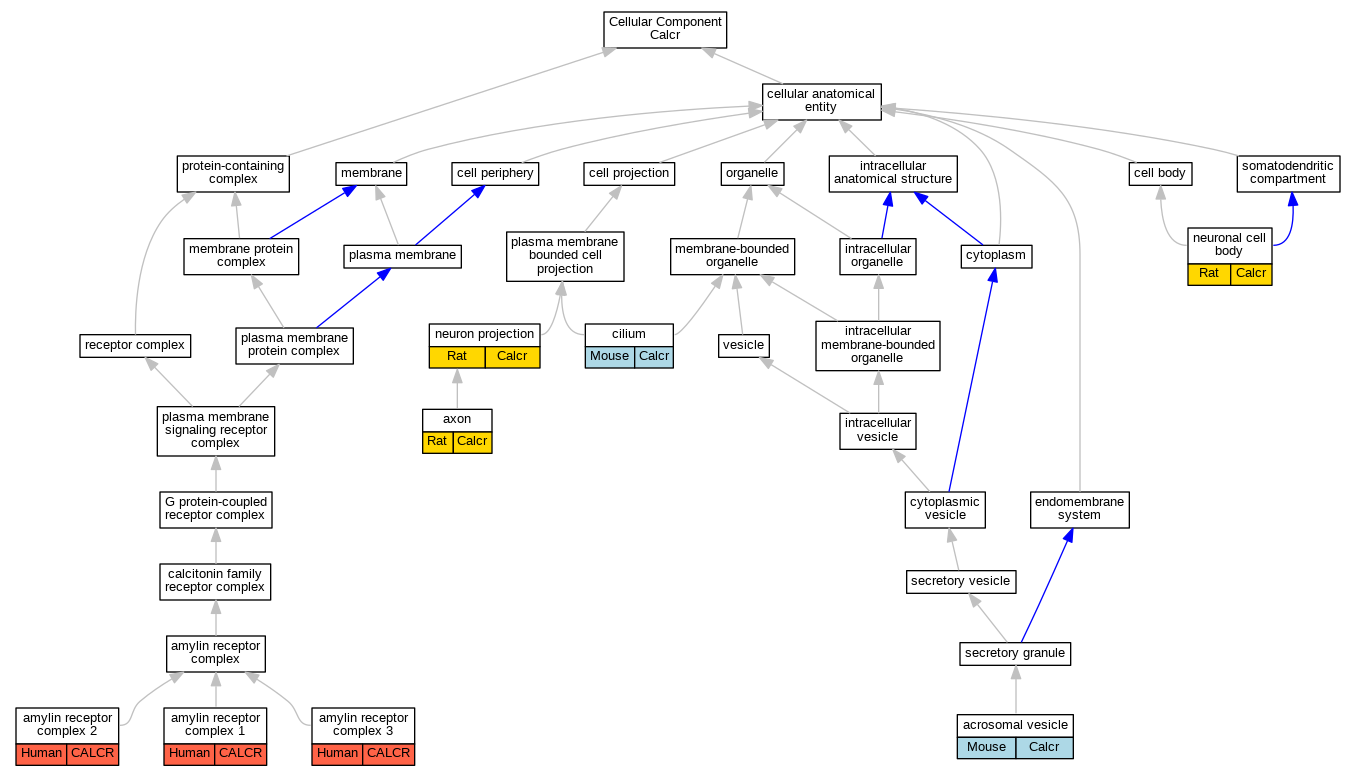
| Cellular Component | GO:0001669 | acrosomal vesicle | Calcr | IDA | | Mouse | PMID:12704735 |
| Cellular Component | GO:0150056 | amylin receptor complex 1 | CALCR | IDA | | Human | PMID:22500019 |
| Cellular Component | GO:0150057 | amylin receptor complex 2 | CALCR | IDA | | Human | PMID:22500019 |
| Cellular Component | GO:0150058 | amylin receptor complex 3 | CALCR | IDA | | Human | PMID:22500019 |
| Cellular Component | GO:0030424 | axon | Calcr | IDA | | Rat | PMID:15571671|RGD:1598634 |
| Cellular Component | GO:0005929 | cilium | Calcr | IDA | | Mouse | PMID:12704735 |
| Cellular Component | GO:0043005 | neuron projection | Calcr | IDA | | Rat | PMID:15571671|RGD:1598634 |
| Cellular Component | GO:0043025 | neuronal cell body | Calcr | IDA | | Rat | PMID:15571671|RGD:1598634 |
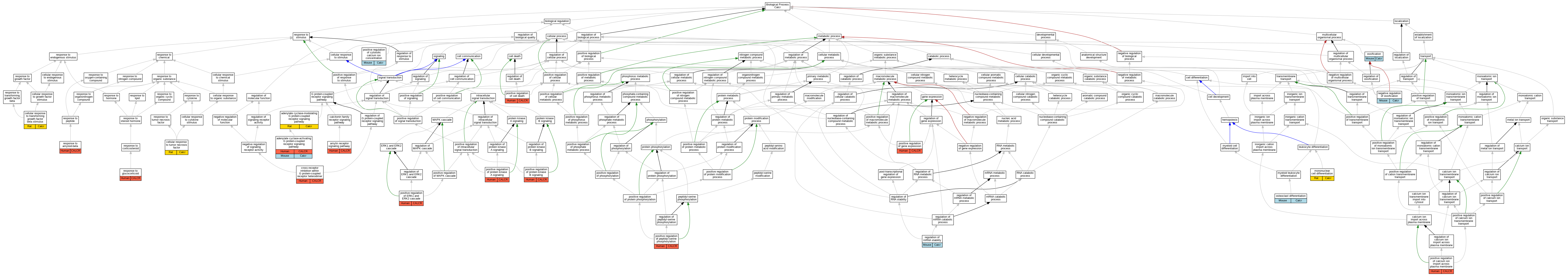
| Biological Process | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | CALCR | IDA | | Human | PMID:14722252 |
| Biological Process | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | CALCR | IDA | | Human | PMID:15563840 |
| Biological Process | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | CALCR | IGI | ComplexPortal:CPX-1134,UniProtKB:O60896 | Human | PMID:22500019 |
| Biological Process | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | CALCR | IGI | UniProtKB:O60894,UniProtKB:P06881 | Human | PMID:22500019 |
| Biological Process | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | CALCR | IGI | UniProtKB:O60894,UniProtKB:P10997:PRO_0000004106 | Human | PMID:22500019 |
| Biological Process | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | CALCR | IGI | UniProtKB:O60894,UniProtKB:P12969:PRO_0000004122 | Human | PMID:22500019 |
| Biological Process | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | CALCR | IGI | UniProtKB:O60894,UniProtKB:P35318 | Human | PMID:22500019 |
| Biological Process | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | CALCR | IGI | UniProtKB:O60895,UniProtKB:P12969:PRO_0000004122 | Human | PMID:22500019 |
| Biological Process | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | CALCR | IGI | UniProtKB:O60895,UniProtKB:P35318 | Human | PMID:22500019 |
| Biological Process | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | CALCR | IGI | UniProtKB:O60896,UniProtKB:P10997:PRO_0000004106 | Human | PMID:22500019 |
| Biological Process | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | CALCR | IGI | UniProtKB:O60896,UniProtKB:P12969:PRO_0000004122 | Human | PMID:22500019 |
| Biological Process | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | CALCR | IDA | | Human | PMID:7619207 |
| Biological Process | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | Calcr | IDA | | Mouse | PMID:7988453 |
| Biological Process | GO:0007188 | adenylate cyclase-modulating G protein-coupled receptor signaling pathway | Calcr | IDA | | Rat | PMID:7723741|RGD:2303052 |
| Biological Process | GO:0097647 | amylin receptor signaling pathway | CALCR | IDA | | Human | PMID:14722252 |
| Biological Process | GO:0097647 | amylin receptor signaling pathway | CALCR | IGI | ComplexPortal:CPX-1134,UniProtKB:O60896 | Human | PMID:22500019 |
| Biological Process | GO:0097647 | amylin receptor signaling pathway | CALCR | IGI | ComplexPortal:CPX-2173 | Human | PMID:22500019 |
| Biological Process | GO:0097647 | amylin receptor signaling pathway | CALCR | IGI | UniProtKB:O60894,UniProtKB:P06881 | Human | PMID:22500019 |
| Biological Process | GO:0097647 | amylin receptor signaling pathway | CALCR | IGI | UniProtKB:O60894,UniProtKB:P10997:PRO_0000004106 | Human | PMID:22500019 |
| Biological Process | GO:0097647 | amylin receptor signaling pathway | CALCR | IGI | UniProtKB:O60894,UniProtKB:P12969:PRO_0000004122 | Human | PMID:22500019 |
| Biological Process | GO:0097647 | amylin receptor signaling pathway | CALCR | IGI | UniProtKB:O60894,UniProtKB:P35318 | Human | PMID:22500019 |
| Biological Process | GO:0097647 | amylin receptor signaling pathway | CALCR | IGI | UniProtKB:O60895,UniProtKB:P12969:PRO_0000004122 | Human | PMID:22500019 |
| Biological Process | GO:0097647 | amylin receptor signaling pathway | CALCR | IGI | UniProtKB:O60895,UniProtKB:P35318 | Human | PMID:22500019 |
| Biological Process | GO:0097647 | amylin receptor signaling pathway | CALCR | IGI | UniProtKB:O60896,UniProtKB:P10997:PRO_0000004106 | Human | PMID:22500019 |
| Biological Process | GO:0071560 | cellular response to transforming growth factor beta stimulus | Calcr | IEP | | Rat | PMID:23110133|RGD:151665477 |
| Biological Process | GO:0071356 | cellular response to tumor necrosis factor | Calcr | IEP | | Rat | PMID:23110133|RGD:151665477 |
| Biological Process | GO:0038041 | cross-receptor inhibition within G protein-coupled receptor heterodimer | CALCR | IPI | UniProtKB:O60896 | Human | PMID:14722252 |
| Biological Process | GO:1903131 | mononuclear cell differentiation | Calcr | IEP | | Rat | PMID:23110133|RGD:151665477 |
| Biological Process | GO:0030279 | negative regulation of ossification | Calcr | IMP | MGI:MGI:3038975 | Mouse | PMID:14970190 |
| Biological Process | GO:0001503 | ossification | Calcr | IMP | MGI:MGI:3038975 | Mouse | PMID:14970190 |
| Biological Process | GO:0030316 | osteoclast differentiation | Calcr | IDA | | Mouse | PMID:7664679 |
| Biological Process | GO:1905665 | positive regulation of calcium ion import across plasma membrane | CALCR | IGI | ComplexPortal:CPX-1134,UniProtKB:O60896 | Human | PMID:22500019 |
| Biological Process | GO:1905665 | positive regulation of calcium ion import across plasma membrane | CALCR | IGI | UniProtKB:O60896,UniProtKB:P10997:PRO_0000004106 | Human | PMID:22500019 |
| Biological Process | GO:0010942 | positive regulation of cell death | CALCR | IGI | ComplexPortal:CPX-1134,UniProtKB:O60896 | Human | PMID:22500019 |
| Biological Process | GO:0010942 | positive regulation of cell death | CALCR | IGI | UniProtKB:O60896,UniProtKB:P10997:PRO_0000004106 | Human | PMID:22500019 |
| Biological Process | GO:0007204 | positive regulation of cytosolic calcium ion concentration | Calcr | IDA | | Mouse | PMID:9753683 |
| Biological Process | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | CALCR | IGI | UniProtKB:O60896,UniProtKB:P05067:PRO_0000000092 | Human | PMID:22500019 |
| Biological Process | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | CALCR | IGI | UniProtKB:O60896,UniProtKB:P10997:PRO_0000004106 | Human | PMID:22500019 |
| Biological Process | GO:0010628 | positive regulation of gene expression | CALCR | IGI | UniProtKB:O60896,UniProtKB:P10997:PRO_0000004106 | Human | PMID:22500019 |
| Biological Process | GO:0033138 | positive regulation of peptidyl-serine phosphorylation | CALCR | IGI | ComplexPortal:CPX-1134,UniProtKB:O60896 | Human | PMID:22500019 |
| Biological Process | GO:0033138 | positive regulation of peptidyl-serine phosphorylation | CALCR | IGI | UniProtKB:O60896,UniProtKB:P10997:PRO_0000004106 | Human | PMID:22500019 |
| Biological Process | GO:0010739 | positive regulation of protein kinase A signaling | CALCR | IGI | UniProtKB:O60896,UniProtKB:P10997:PRO_0000004106 | Human | PMID:22500019 |
| Biological Process | GO:0051897 | positive regulation of protein kinase B signaling | CALCR | IGI | ComplexPortal:CPX-1134,UniProtKB:O60896 | Human | PMID:22500019 |
| Biological Process | GO:0051897 | positive regulation of protein kinase B signaling | CALCR | IGI | UniProtKB:O60896,UniProtKB:P10997:PRO_0000004106 | Human | PMID:22500019 |
| Biological Process | GO:0043488 | regulation of mRNA stability | Calcr | IDA | | Mouse | PMID:9002981 |
| Biological Process | GO:1904645 | response to amyloid-beta | CALCR | IGI | UniProtKB:O60896 | Human | PMID:22500019 |
| Biological Process | GO:0051384 | response to glucocorticoid | CALCR | IDA | | Human | PMID:11250927 |
 Analysis Tools
Analysis Tools