|
Source |
Alliance of Genome Resources |
| Category | ID | Classification term | Gene | Evidence | Inferred from | Organism | Reference |
|---|---|---|---|---|---|---|---|
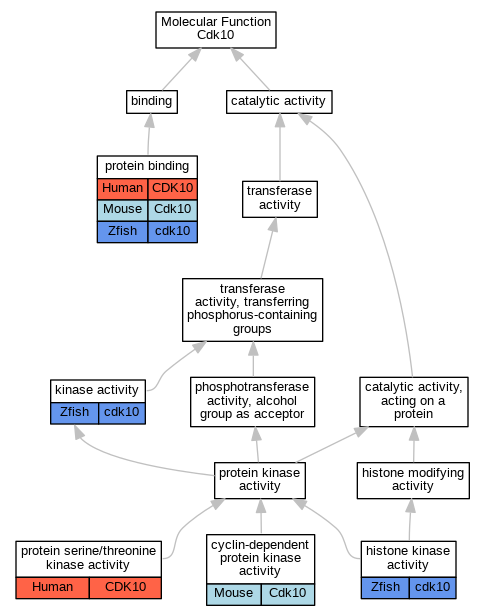
| Molecular Function | GO:0097472 | cyclin-dependent protein kinase activity | Cdk10 | IGI | MGI:MGI:1916359 | Mouse | PMID:24218572 |
| Molecular Function | GO:0035173 | histone kinase activity | cdk10 | IDA | Zfish | PMID:23902762 | |
| Molecular Function | GO:0016301 | kinase activity | cdk10 | IDA | Zfish | PMID:23902762 | |
| Molecular Function | GO:0005515 | protein binding | CDK10 | IPI | UniProtKB:P15036 | Human | PMID:18242510 |
| Molecular Function | GO:0005515 | protein binding | CDK10 | IPI | UniProtKB:Q13526 | Human | PMID:22158035 |
| Molecular Function | GO:0005515 | protein binding | CDK10 | IPI | UniProtKB:P08238 | Human | PMID:22939624 |
| Molecular Function | GO:0005515 | protein binding | CDK10 | IPI | UniProtKB:Q16513 | Human | PMID:27104747 |
| Molecular Function | GO:0005515 | protein binding | CDK10 | IPI | UniProtKB:P08238 | Human | PMID:32707033 |
| Molecular Function | GO:0005515 | protein binding | Cdk10 | IPI | PR:Q8QZR8 | Mouse | PMID:24218572 |
| Molecular Function | GO:0005515 | protein binding | cdk10 | IPI | RefSeq:NM_001256295 | Zfish | PMID:23902762 |
| Molecular Function | GO:0004674 | protein serine/threonine kinase activity | CDK10 | IDA | Human | PMID:27104747 | |
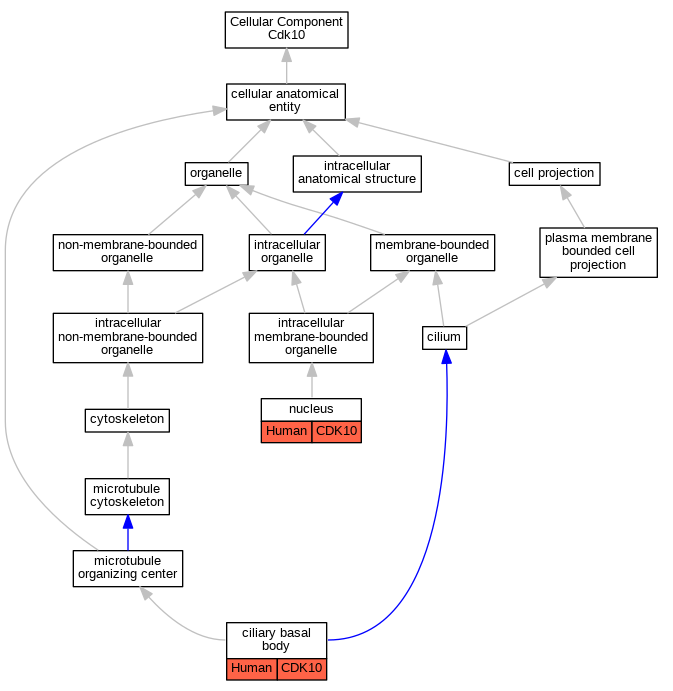
| Cellular Component | GO:0036064 | ciliary basal body | CDK10 | IDA | Human | PMID:27104747 | |
| Cellular Component | GO:0005634 | nucleus | CDK10 | IDA | Human | PMID:11006117 | |
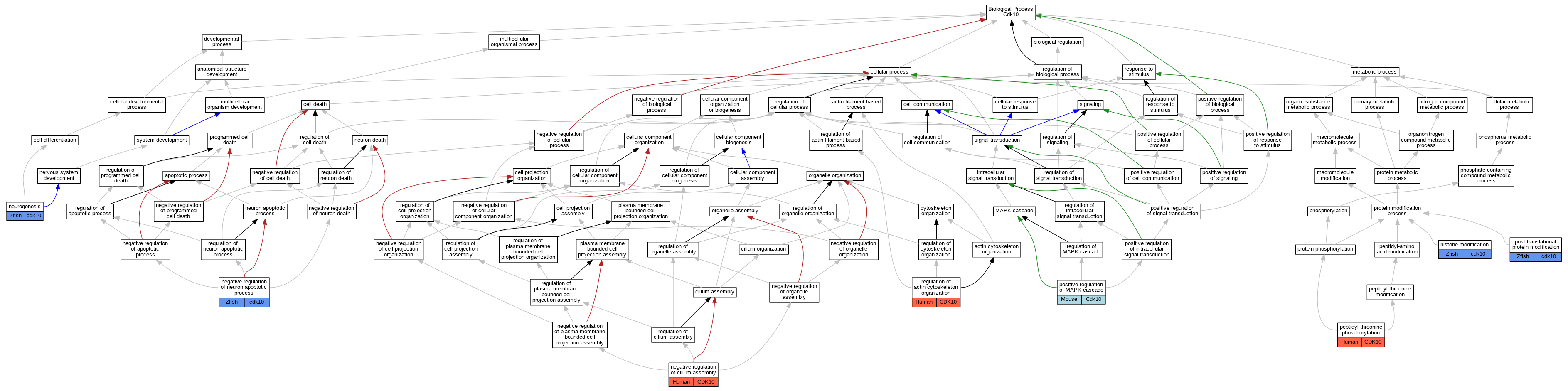
| Biological Process | GO:0016570 | histone modification | cdk10 | IDA | Zfish | PMID:23902762|ZFIN:ZDB-PUB-130816-36 | |
| Biological Process | GO:1902018 | negative regulation of cilium assembly | CDK10 | IMP | Human | PMID:27104747 | |
| Biological Process | GO:0043524 | negative regulation of neuron apoptotic process | cdk10 | IMP | ZFIN:ZDB-MRPHLNO-131126-4 | Zfish | PMID:23902762 |
| Biological Process | GO:0022008 | neurogenesis | cdk10 | IMP | ZFIN:ZDB-MRPHLNO-131126-4 | Zfish | PMID:23902762 |
| Biological Process | GO:0018107 | peptidyl-threonine phosphorylation | CDK10 | IDA | Human | PMID:27104747 | |
| Biological Process | GO:0043410 | positive regulation of MAPK cascade | Cdk10 | IGI | MGI:MGI:1916359 | Mouse | PMID:24218572 |
| Biological Process | GO:0043687 | post-translational protein modification | cdk10 | IDA | Zfish | PMID:23902762 | |
| Biological Process | GO:0032956 | regulation of actin cytoskeleton organization | CDK10 | IMP | Human | PMID:27104747 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 08/02/2024 MGI 6.24 |

|
|
|
||