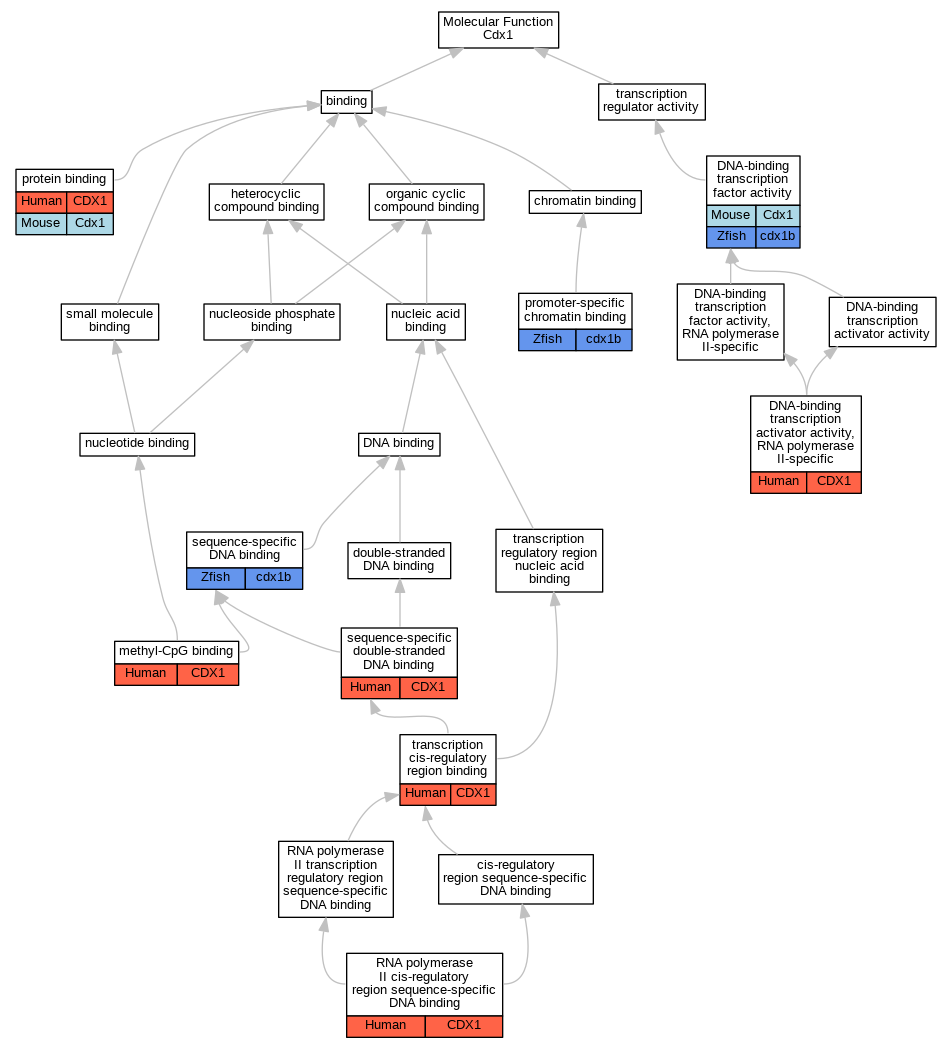
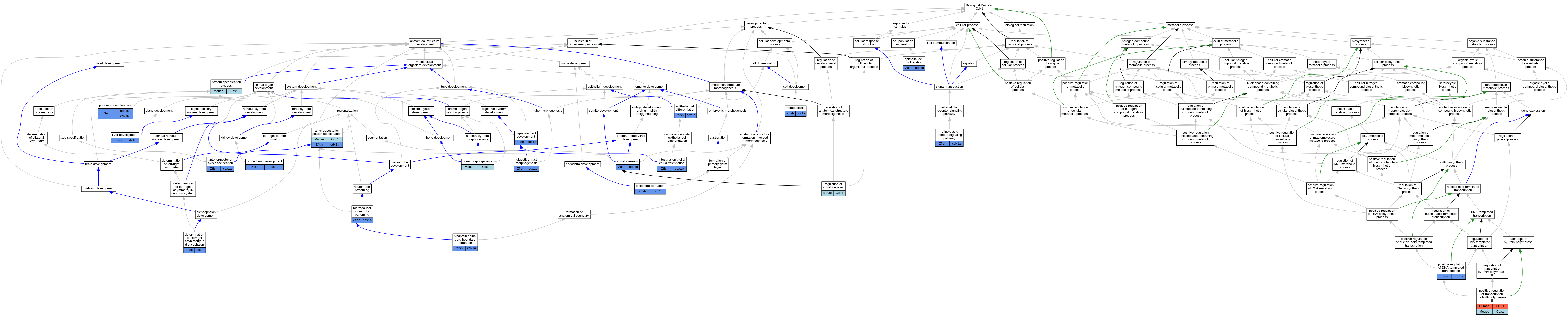
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0001228 | DNA-binding transcription activator activity, RNA polymerase II-specific | CDX1 | IDA | | Human | PMID:15774940 |
| Molecular Function | GO:0003700 | DNA-binding transcription factor activity | Cdx1 | IDA | | Mouse | MGI:MGI:5301472|PMID:22015720 |
| Molecular Function | GO:0003700 | DNA-binding transcription factor activity | cdx1b | IPI | ZFIN:ZDB-GENE-980526-404 | Zfish | PMID:18234726 |
| Molecular Function | GO:0008327 | methyl-CpG binding | CDX1 | IDA | | Human | PMID:28473536 |
| Molecular Function | GO:1990841 | promoter-specific chromatin binding | cdx1b | IDA | | Zfish | PMID:33197427 |
| Molecular Function | GO:0005515 | protein binding | CDX1 | IPI | UniProtKB:P49715 | Human | PMID:19059241 |
| Molecular Function | GO:0005515 | protein binding | Cdx1 | IPI | UniProtKB:P20226 | Mouse | MGI:MGI:6294231|PMID:17158164 |
| Molecular Function | GO:0000978 | RNA polymerase II cis-regulatory region sequence-specific DNA binding | CDX1 | IDA | | Human | PMID:15774940 |
| Molecular Function | GO:0043565 | sequence-specific DNA binding | cdx1b | IPI | ZFIN:ZDB-GENE-980526-404 | Zfish | PMID:18234726 |
| Molecular Function | GO:1990837 | sequence-specific double-stranded DNA binding | CDX1 | IDA | | Human | PMID:28473536 |
| Molecular Function | GO:0000976 | transcription cis-regulatory region binding | CDX1 | IDA | | Human | PMID:24623306 |
| Biological Process | GO:0009948 | anterior/posterior axis specification | cdx1a | IGI | ZFIN:ZDB-GENE-000823-1,ZFIN:ZDB-GENO-980202-1126,ZFIN:ZDB-MRPHLNO-060602-1 | Zfish | PMID:16457800 |
| Biological Process | GO:0009948 | anterior/posterior axis specification | cdx1a | IGI | ZFIN:ZDB-GENE-000329-2,ZFIN:ZDB-GENO-980202-1126,ZFIN:ZDB-MRPHLNO-060602-1 | Zfish | PMID:16457800 |
| Biological Process | GO:0009952 | anterior/posterior pattern specification | Cdx1 | IMP | MGI:MGI:2154514 | Mouse | PMID:11784046 |
| Biological Process | GO:0009952 | anterior/posterior pattern specification | Cdx1 | IGI | MGI:MGI:97858 | Mouse | PMID:11784046 |
| Biological Process | GO:0009952 | anterior/posterior pattern specification | Cdx1 | IMP | MGI:MGI:2154514 | Mouse | PMID:7585967 |
| Biological Process | GO:0009952 | anterior/posterior pattern specification | cdx1a | IGI | ZFIN:ZDB-GENO-080715-2,ZFIN:ZDB-MRPHLNO-050509-3 | Zfish | PMID:18234725 |
| Biological Process | GO:0009952 | anterior/posterior pattern specification | cdx1a | IGI | ZFIN:ZDB-GENO-080215-1,ZFIN:ZDB-MRPHLNO-060602-1 | Zfish | PMID:17953490 |
| Biological Process | GO:0060349 | bone morphogenesis | Cdx1 | IGI | MGI:MGI:97858 | Mouse | PMID:11784046 |
| Biological Process | GO:0060349 | bone morphogenesis | Cdx1 | IMP | MGI:MGI:2154514 | Mouse | PMID:11784046 |
| Biological Process | GO:0035462 | determination of left/right asymmetry in diencephalon | cdx1b | IMP | ZFIN:ZDB-GENO-220527-3 | Zfish | PMID:33197427 |
| Biological Process | GO:0048565 | digestive tract development | cdx1b | IMP | ZFIN:ZDB-MRPHLNO-080707-1 | Zfish | PMID:19253392 |
| Biological Process | GO:0048546 | digestive tract morphogenesis | cdx1b | IMP | ZFIN:ZDB-MRPHLNO-090217-1 | Zfish | PMID:18804112 |
| Biological Process | GO:0048546 | digestive tract morphogenesis | cdx1b | IMP | ZFIN:ZDB-MRPHLNO-090217-2 | Zfish | PMID:18804112 |
| Biological Process | GO:0048546 | digestive tract morphogenesis | cdx1b | IMP | ZFIN:ZDB-MRPHLNO-080707-1 | Zfish | PMID:18234726 |
| Biological Process | GO:0001706 | endoderm formation | cdx1b | IMP | ZFIN:ZDB-MRPHLNO-080707-1 | Zfish | PMID:18234726 |
| Biological Process | GO:0030855 | epithelial cell differentiation | cdx1b | IMP | ZFIN:ZDB-MRPHLNO-090217-1 | Zfish | PMID:18804112 |
| Biological Process | GO:0050673 | epithelial cell proliferation | cdx1b | IMP | ZFIN:ZDB-MRPHLNO-090217-1 | Zfish | PMID:18804112 |
| Biological Process | GO:0030097 | hemopoiesis | cdx1a | IGI | ZFIN:ZDB-GENO-980202-1126,ZFIN:ZDB-MRPHLNO-060602-1 | Zfish | PMID:16457800 |
| Biological Process | GO:0021906 | hindbrain-spinal cord boundary formation | cdx1a | IMP | ZFIN:ZDB-MRPHLNO-050509-3 | Zfish | PMID:17507415 |
| Biological Process | GO:0060575 | intestinal epithelial cell differentiation | cdx1b | IMP | ZFIN:ZDB-MRPHLNO-080707-1 | Zfish | PMID:19253392 |
| Biological Process | GO:0001889 | liver development | cdx1b | IMP | ZFIN:ZDB-MRPHLNO-080707-1 | Zfish | PMID:18234726 |
| Biological Process | GO:0031016 | pancreas development | cdx1a | IGI | ZFIN:ZDB-GENO-080715-2,ZFIN:ZDB-MRPHLNO-050509-3 | Zfish | PMID:18234725 |
| Biological Process | GO:0031016 | pancreas development | cdx1b | IMP | ZFIN:ZDB-MRPHLNO-080707-1 | Zfish | PMID:18234726 |
| Biological Process | GO:0007389 | pattern specification process | Cdx1 | IMP | MGI:MGI:2154514 | Mouse | PMID:11959827 |
| Biological Process | GO:0045893 | positive regulation of DNA-templated transcription | cdx1b | IGI | ZFIN:ZDB-GENE-980526-404 | Zfish | PMID:18234726 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | CDX1 | IDA | | Human | PMID:15774940 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | CDX1 | IMP | | Human | PMID:24623306 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | Cdx1 | IDA | | Mouse | PMID:7585967 |
| Biological Process | GO:0048793 | pronephros development | cdx1a | IGI | ZFIN:ZDB-GENO-080215-1,ZFIN:ZDB-MRPHLNO-060602-1 | Zfish | PMID:17953490 |
| Biological Process | GO:0014807 | regulation of somitogenesis | Cdx1 | IMP | | Mouse | MGI:MGI:5301472|PMID:22015720 |
| Biological Process | GO:0048384 | retinoic acid receptor signaling pathway | cdx1a | IGI | ZFIN:ZDB-GENO-080215-1,ZFIN:ZDB-MRPHLNO-060602-1 | Zfish | PMID:17953490 |
| Biological Process | GO:0021903 | rostrocaudal neural tube patterning | cdx1a | IGI | ZFIN:ZDB-MRPHLNO-050509-3,ZFIN:ZDB-MRPHLNO-050509-7 | Zfish | PMID:17079270 |
| Biological Process | GO:0001756 | somitogenesis | cdx1a | IGI | ZFIN:ZDB-MRPHLNO-050509-7 | Zfish | PMID:15708563 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
 Analysis Tools
Analysis Tools