| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0005095 | GTPase inhibitor activity | Cpeb2 | IDA | | Rat | PMID:22157746|RGD:8553654 |
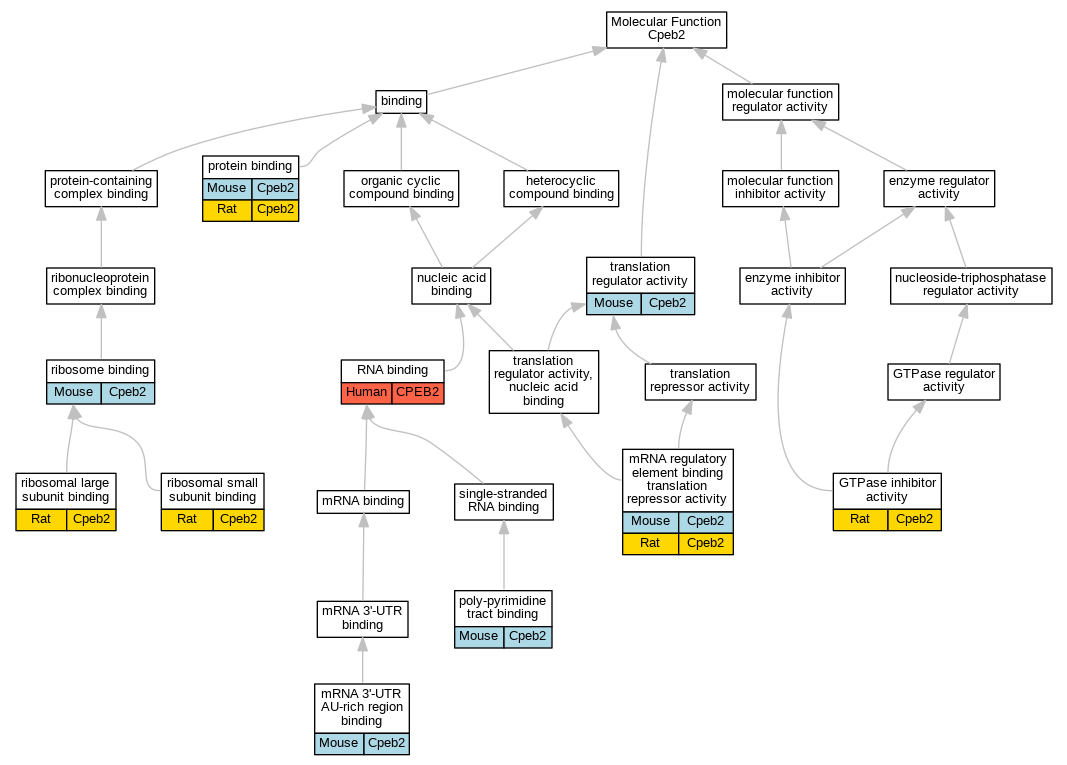
| Molecular Function | GO:0035925 | mRNA 3'-UTR AU-rich region binding | Cpeb2 | IDA | | Mouse | MGI:MGI:5543770|PMID:18752464 |
| Molecular Function | GO:0000900 | mRNA regulatory element binding translation repressor activity | Cpeb2 | IDA | | Mouse | MGI:MGI:5543770|PMID:18752464 |
| Molecular Function | GO:0000900 | mRNA regulatory element binding translation repressor activity | Cpeb2 | IDA | | Rat | PMID:22157746|RGD:8553654 |
| Molecular Function | GO:0008187 | poly-pyrimidine tract binding | Cpeb2 | IDA | | Mouse | PMID:12672660 |
| Molecular Function | GO:0005515 | protein binding | Cpeb2 | IPI | PR:Q91YI6 | Mouse | PMID:17927953 |
| Molecular Function | GO:0005515 | protein binding | Cpeb2 | IPI | PR:P58252 | Mouse | PMID:22157746 |
| Molecular Function | GO:0005515 | protein binding | Cpeb2 | IPI | UniProtKB:P58252 | Rat | PMID:22157746|RGD:8553654 |
| Molecular Function | GO:0043023 | ribosomal large subunit binding | Cpeb2 | IDA | | Rat | PMID:22157746|RGD:8553654 |
| Molecular Function | GO:0043024 | ribosomal small subunit binding | Cpeb2 | IDA | | Rat | PMID:22157746|RGD:8553654 |
| Molecular Function | GO:0043022 | ribosome binding | Cpeb2 | IDA | | Mouse | MGI:MGI:5314154|PMID:22157746 |
| Molecular Function | GO:0003723 | RNA binding | CPEB2 | HDA | | Human | PMID:22658674 |
| Molecular Function | GO:0003723 | RNA binding | CPEB2 | HDA | | Human | PMID:22681889 |
| Molecular Function | GO:0045182 | translation regulator activity | Cpeb2 | IDA | | Mouse | PMID:29141213 |
| Molecular Function | GO:0045182 | translation regulator activity | Cpeb2 | IMP | | Mouse | PMID:29141213 |
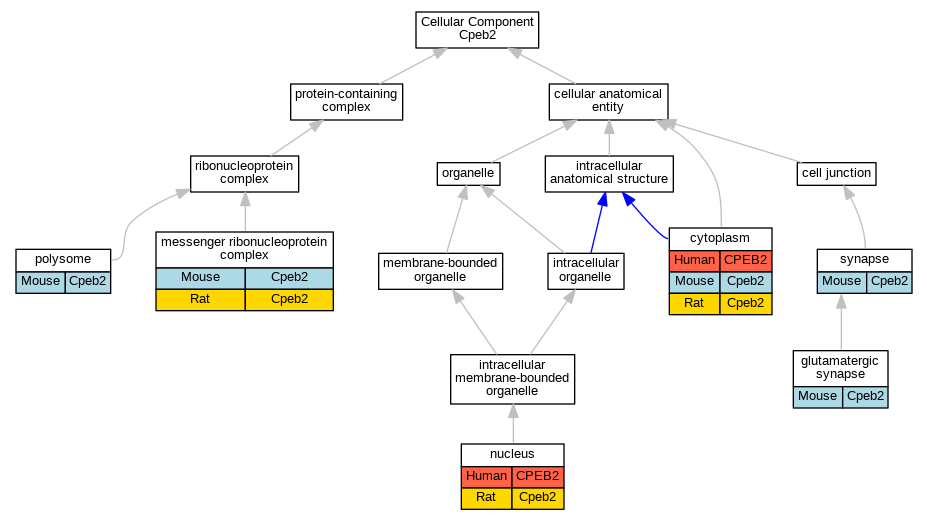
| Cellular Component | GO:0005737 | cytoplasm | CPEB2 | IDA | | Human | PMID:20639532 |
| Cellular Component | GO:0005737 | cytoplasm | Cpeb2 | IDA | | Mouse | PMID:12672660 |
| Cellular Component | GO:0005737 | cytoplasm | Cpeb2 | IDA | | Rat | PMID:22157746|RGD:8553654 |
| Cellular Component | GO:0098978 | glutamatergic synapse | Cpeb2 | IMP | | Mouse | PMID:29141213 |
| Cellular Component | GO:0098978 | glutamatergic synapse | Cpeb2 | IDA | | Mouse | PMID:29141213 |
| Cellular Component | GO:1990124 | messenger ribonucleoprotein complex | Cpeb2 | IDA | | Mouse | MGI:MGI:5314154|PMID:22157746 |
| Cellular Component | GO:1990124 | messenger ribonucleoprotein complex | Cpeb2 | IDA | | Rat | PMID:22157746|RGD:8553654 |
| Cellular Component | GO:0005634 | nucleus | CPEB2 | IDA | | Human | PMID:20639532 |
| Cellular Component | GO:0005634 | nucleus | Cpeb2 | IDA | | Rat | PMID:22157746|RGD:8553654 |
| Cellular Component | GO:0005844 | polysome | Cpeb2 | IDA | | Mouse | PMID:22157746 |
| Cellular Component | GO:0045202 | synapse | Cpeb2 | IMP | | Mouse | MGI:MGI:6104123|PMID:29141213 |
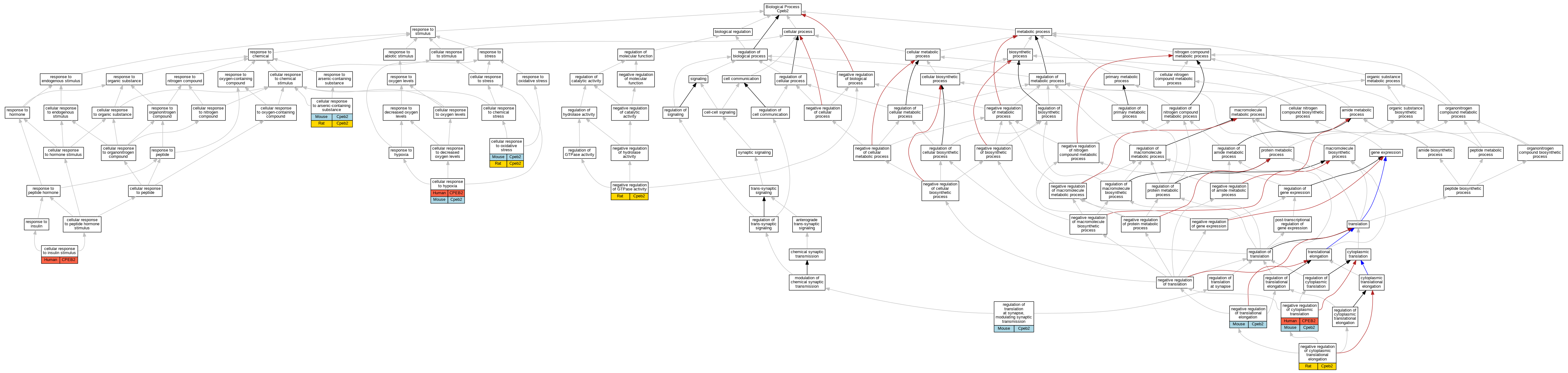
| Biological Process | GO:0071243 | cellular response to arsenic-containing substance | Cpeb2 | IDA | | Mouse | MGI:MGI:5314154|PMID:22157746 |
| Biological Process | GO:0071243 | cellular response to arsenic-containing substance | Cpeb2 | IDA | | Rat | PMID:22157746|RGD:8553654 |
| Biological Process | GO:0071456 | cellular response to hypoxia | CPEB2 | IDA | | Human | PMID:18752464 |
| Biological Process | GO:0071456 | cellular response to hypoxia | Cpeb2 | IDA | | Mouse | MGI:MGI:5543770|PMID:18752464 |
| Biological Process | GO:0032869 | cellular response to insulin stimulus | CPEB2 | IDA | | Human | PMID:18752464 |
| Biological Process | GO:0034599 | cellular response to oxidative stress | Cpeb2 | IDA | | Mouse | MGI:MGI:5314154|PMID:22157746 |
| Biological Process | GO:0034599 | cellular response to oxidative stress | Cpeb2 | IDA | | Rat | PMID:22157746|RGD:8553654 |
| Biological Process | GO:2000766 | negative regulation of cytoplasmic translation | CPEB2 | IMP | | Human | PMID:18752464 |
| Biological Process | GO:2000766 | negative regulation of cytoplasmic translation | Cpeb2 | IDA | | Mouse | MGI:MGI:5543770|PMID:18752464 |
| Biological Process | GO:1900248 | negative regulation of cytoplasmic translational elongation | Cpeb2 | IDA | | Rat | PMID:22157746|RGD:8553654 |
| Biological Process | GO:0034260 | negative regulation of GTPase activity | Cpeb2 | IDA | | Rat | PMID:22157746|RGD:8553654 |
| Biological Process | GO:0045900 | negative regulation of translational elongation | Cpeb2 | IGI | MGI:MGI:95288 | Mouse | PMID:22157746 |
| Biological Process | GO:0099547 | regulation of translation at synapse, modulating synaptic transmission | Cpeb2 | IDA | | Mouse | PMID:29141213 |
| Biological Process | GO:0099547 | regulation of translation at synapse, modulating synaptic transmission | Cpeb2 | IMP | | Mouse | PMID:29141213 |
 Analysis Tools
Analysis Tools