|
Source |
Alliance of Genome Resources |
| Category | ID | Classification term | Gene | Evidence | Inferred from | Organism | Reference |
|---|---|---|---|---|---|---|---|
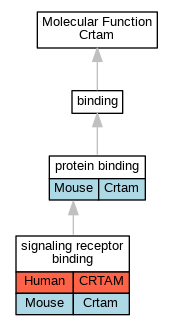
| Molecular Function | GO:0005515 | protein binding | Crtam | IPI | UniProtKB:Q80U72 | Mouse | MGI:MGI:3834299|PMID:18329370 |
| Molecular Function | GO:0005102 | signaling receptor binding | CRTAM | IPI | UniProtKB:Q9BY67 | Human | PMID:15811952 |
| Molecular Function | GO:0005102 | signaling receptor binding | Crtam | IPI | UniProtKB:Q8R5M8 | Mouse | MGI:MGI:3689781|PMID:15811952 |
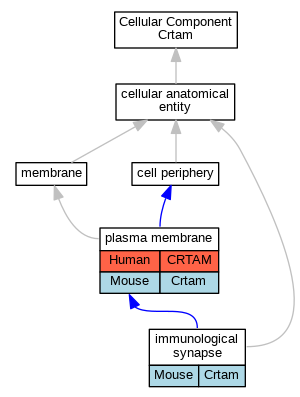
| Cellular Component | GO:0001772 | immunological synapse | Crtam | IDA | Mouse | MGI:MGI:3834299|PMID:18329370 | |
| Cellular Component | GO:0005886 | plasma membrane | CRTAM | IDA | Human | PMID:15811952 | |
| Cellular Component | GO:0005886 | plasma membrane | Crtam | IDA | Mouse | MGI:MGI:5581366|PMID:24687959 | |
| Cellular Component | GO:0005886 | plasma membrane | Crtam | IDA | Mouse | MGI:MGI:3689781|PMID:15811952 | |
| Cellular Component | GO:0005886 | plasma membrane | Crtam | IDA | Mouse | MGI:MGI:4359960|PMID:19752223 | |
| Cellular Component | GO:0005886 | plasma membrane | Crtam | IDA | Mouse | MGI:MGI:3834299|PMID:18329370 | |
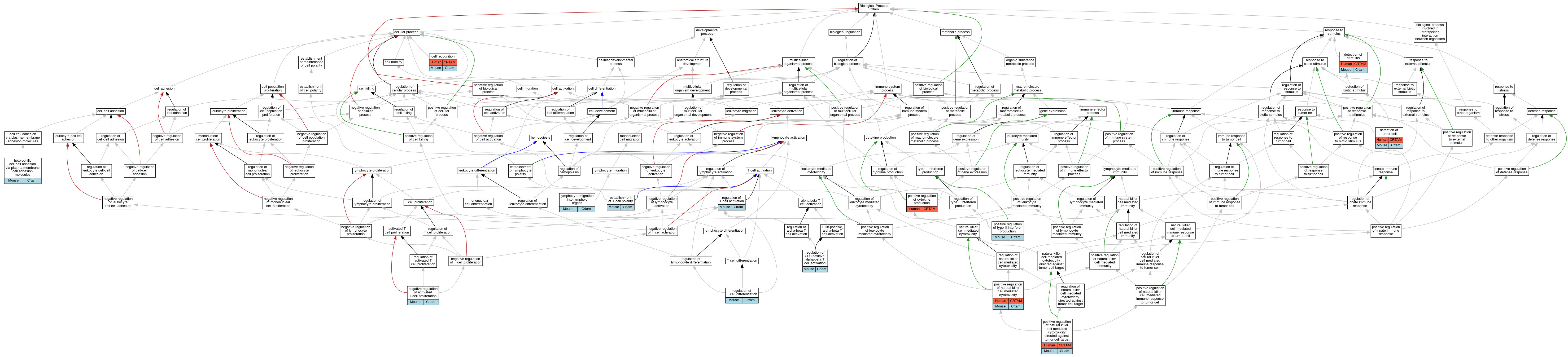
| Biological Process | GO:0008037 | cell recognition | CRTAM | IDA | Human | PMID:15811952 | |
| Biological Process | GO:0008037 | cell recognition | Crtam | IDA | Mouse | MGI:MGI:3689781|PMID:15811952 | |
| Biological Process | GO:0051606 | detection of stimulus | CRTAM | IDA | Human | PMID:15811952 | |
| Biological Process | GO:0051606 | detection of stimulus | Crtam | IDA | Mouse | MGI:MGI:3689781|PMID:15811952 | |
| Biological Process | GO:0002355 | detection of tumor cell | CRTAM | IDA | Human | PMID:15811952 | |
| Biological Process | GO:0002355 | detection of tumor cell | Crtam | IDA | Mouse | MGI:MGI:3689781|PMID:15811952 | |
| Biological Process | GO:0001768 | establishment of T cell polarity | Crtam | IDA | Mouse | MGI:MGI:3834299|PMID:18329370 | |
| Biological Process | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules | Crtam | IMP | Mouse | MGI:MGI:4359960|PMID:19752223 | |
| Biological Process | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules | Crtam | IMP | Mouse | MGI:MGI:5581366|PMID:24687959 | |
| Biological Process | GO:0097021 | lymphocyte migration into lymphoid organs | Crtam | IMP | Mouse | MGI:MGI:5581366|PMID:24687959 | |
| Biological Process | GO:0046007 | negative regulation of activated T cell proliferation | Crtam | IMP | Mouse | MGI:MGI:3834299|PMID:18329370 | |
| Biological Process | GO:0001819 | positive regulation of cytokine production | CRTAM | IDA | Human | PMID:15811952 | |
| Biological Process | GO:0045954 | positive regulation of natural killer cell mediated cytotoxicity | CRTAM | IDA | Human | PMID:15811952 | |
| Biological Process | GO:0045954 | positive regulation of natural killer cell mediated cytotoxicity | Crtam | IDA | Mouse | MGI:MGI:3689781|PMID:15811952 | |
| Biological Process | GO:0002860 | positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target | CRTAM | IDA | Human | PMID:15811952 | |
| Biological Process | GO:0002860 | positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target | Crtam | IDA | Mouse | MGI:MGI:3689781|PMID:15811952 | |
| Biological Process | GO:0032729 | positive regulation of type II interferon production | Crtam | IMP | Mouse | MGI:MGI:3834299|PMID:18329370 | |
| Biological Process | GO:2001185 | regulation of CD8-positive, alpha-beta T cell activation | Crtam | IMP | Mouse | MGI:MGI:4359960|PMID:19752223 | |
| Biological Process | GO:0050863 | regulation of T cell activation | Crtam | IMP | Mouse | MGI:MGI:5581366|PMID:24687959 | |
| Biological Process | GO:0045580 | regulation of T cell differentiation | Crtam | IMP | Mouse | MGI:MGI:5581366|PMID:24687959 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 06/12/2024 MGI 6.13 |

|
|
|
||