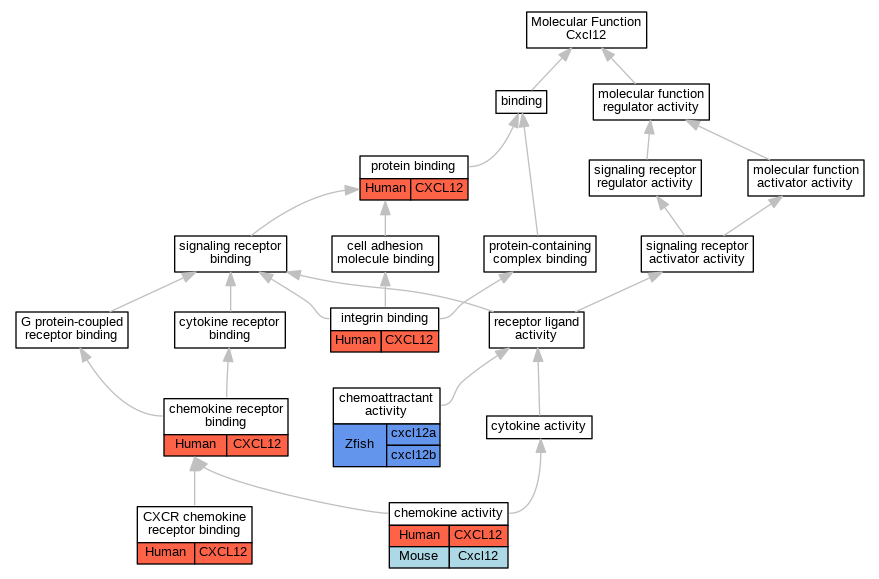
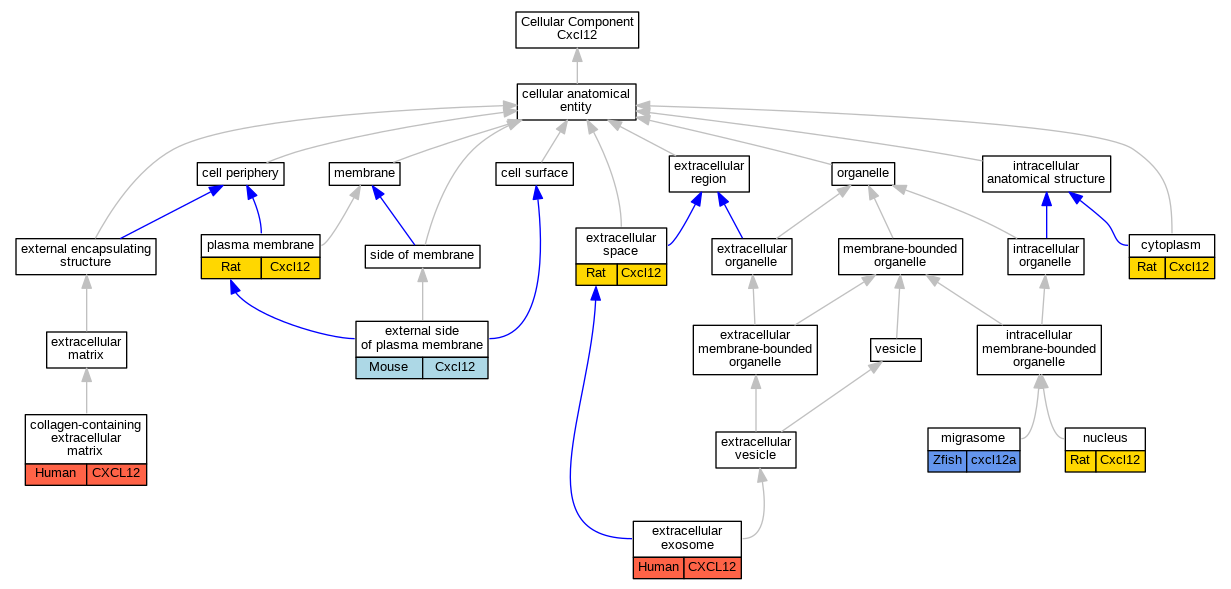
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0042056 | chemoattractant activity | cxcl12a | IMP | ZFIN:ZDB-GENO-200709-9 | Zfish | PMID:31371827 |
| Molecular Function | GO:0042056 | chemoattractant activity | cxcl12a | IMP | | Zfish | PMID:16129886 |
| Molecular Function | GO:0042056 | chemoattractant activity | cxcl12a | IDA | | Zfish | PMID:18579679 |
| Molecular Function | GO:0042056 | chemoattractant activity | cxcl12b | IDA | | Zfish | PMID:18579679 |
| Molecular Function | GO:0008009 | chemokine activity | CXCL12 | IDA | | Human | PMID:18308860 |
| Molecular Function | GO:0008009 | chemokine activity | CXCL12 | IMP | | Human | PMID:27400149 |
| Molecular Function | GO:0008009 | chemokine activity | Cxcl12 | IDA | | Mouse | PMID:12183377 |
| Molecular Function | GO:0042379 | chemokine receptor binding | CXCL12 | IMP | | Human | PMID:22457824 |
| Molecular Function | GO:0045236 | CXCR chemokine receptor binding | CXCL12 | IDA | | Human | PMID:20388803 |
| Molecular Function | GO:0005178 | integrin binding | CXCL12 | IDA | | Human | PMID:29301984 |
| Molecular Function | GO:0005515 | protein binding | CXCL12 | IPI | UniProtKB:O00585 | Human | PMID:28381538 |
| Molecular Function | GO:0005515 | protein binding | CXCL12 | IPI | UniProtKB:O14625 | Human | PMID:28381538 |
| Molecular Function | GO:0005515 | protein binding | CXCL12 | IPI | UniProtKB:O15444 | Human | PMID:28381538 |
| Molecular Function | GO:0005515 | protein binding | CXCL12 | IPI | UniProtKB:O95715 | Human | PMID:28381538 |
| Molecular Function | GO:0005515 | protein binding | CXCL12 | IPI | UniProtKB:P02776 | Human | PMID:28381538 |
| Molecular Function | GO:0005515 | protein binding | CXCL12 | IPI | UniProtKB:P02778 | Human | PMID:28381538 |
| Molecular Function | GO:0005515 | protein binding | CXCL12 | IPI | UniProtKB:P10720 | Human | PMID:28381538 |
| Molecular Function | GO:0005515 | protein binding | CXCL12 | IPI | UniProtKB:P13501 | Human | PMID:28381538 |
| Molecular Function | GO:0005515 | protein binding | CXCL12 | IPI | UniProtKB:P13501:PRO_0000005175 | Human | PMID:28381538 |
| Molecular Function | GO:0005515 | protein binding | CXCL12 | IPI | UniProtKB:P19875 | Human | PMID:28381538 |
| Molecular Function | GO:0005515 | protein binding | CXCL12 | IPI | UniProtKB:P47992 | Human | PMID:28381538 |
| Molecular Function | GO:0005515 | protein binding | CXCL12 | IPI | UniProtKB:P51671 | Human | PMID:28381538 |
| Molecular Function | GO:0005515 | protein binding | CXCL12 | IPI | UniProtKB:P78556 | Human | PMID:28381538 |
| Molecular Function | GO:0005515 | protein binding | CXCL12 | IPI | UniProtKB:P80162 | Human | PMID:28381538 |
| Molecular Function | GO:0005515 | protein binding | CXCL12 | IPI | UniProtKB:Q07325 | Human | PMID:28381538 |
| Molecular Function | GO:0005515 | protein binding | CXCL12 | IPI | UniProtKB:Q6UXB2 | Human | PMID:28381538 |
| Molecular Function | GO:0005515 | protein binding | CXCL12 | IPI | UniProtKB:Q99616 | Human | PMID:28381538 |
| Molecular Function | GO:0005515 | protein binding | CXCL12 | IPI | UniProtKB:Q9NRJ3 | Human | PMID:28381538 |
| Molecular Function | GO:0005515 | protein binding | CXCL12 | IPI | UniProtKB:Q9UBD3 | Human | PMID:28381538 |
| Molecular Function | GO:0005515 | protein binding | CXCL12 | IPI | UniProtKB:Q9Y258 | Human | PMID:28381538 |
| Cellular Component | GO:0062023 | collagen-containing extracellular matrix | CXCL12 | HDA | | Human | PMID:25037231 |
| Cellular Component | GO:0005737 | cytoplasm | Cxcl12 | IDA | | Rat | PMID:24924806|RGD:11352662 |
| Cellular Component | GO:0009897 | external side of plasma membrane | Cxcl12 | IDA | | Mouse | PMID:19008373 |
| Cellular Component | GO:0070062 | extracellular exosome | CXCL12 | HDA | | Human | PMID:23533145 |
| Cellular Component | GO:0005615 | extracellular space | Cxcl12 | IDA | | Rat | PMID:11820456|RGD:632503 |
| Cellular Component | GO:0140494 | migrasome | cxcl12a | IMP | ZFIN:ZDB-GENO-200709-9 | Zfish | PMID:31371827 |
| Cellular Component | GO:0005634 | nucleus | Cxcl12 | IDA | | Rat | PMID:29550796|RGD:13792610 |
| Cellular Component | GO:0005886 | plasma membrane | Cxcl12 | IDA | | Rat | PMID:17046839|RGD:2306583 |
| Biological Process | GO:0008344 | adult locomotory behavior | Cxcl12 | IMP | | Rat | PMID:19303923|RGD:2306565 |
| Biological Process | GO:0009887 | animal organ morphogenesis | cxcl12a | IMP | | Zfish | PMID:11748832 |
| Biological Process | GO:0031100 | animal organ regeneration | Cxcl12 | IMP | | Rat | PMID:17148669|RGD:2306581 |
| Biological Process | GO:0007411 | axon guidance | cxcl12a | IGI | ZFIN:ZDB-MRPHLNO-050318-6,ZFIN:ZDB-MRPHLNO-050318-7 | Zfish | PMID:15716407 |
| Biological Process | GO:0007411 | axon guidance | cxcl12a | IMP | ZFIN:ZDB-MRPHLNO-050324-1 | Zfish | PMID:15659553 |
| Biological Process | GO:0007411 | axon guidance | cxcl12a | IMP | ZFIN:ZDB-MRPHLNO-050324-1 | Zfish | PMID:19595689 |
| Biological Process | GO:0043534 | blood vessel endothelial cell migration | Cxcl12 | IMP | MGI:MGI:1934384 | Mouse | PMID:15626744 |
| Biological Process | GO:0007420 | brain development | Cxcl12 | IDA | | Mouse | PMID:12183377 |
| Biological Process | GO:0007420 | brain development | Cxcl12 | IEP | | Rat | PMID:12401342|RGD:1358280 |
| Biological Process | GO:0001569 | branching involved in blood vessel morphogenesis | Cxcl12 | IMP | MGI:MGI:1934384 | Mouse | PMID:15626744 |
| Biological Process | GO:0060326 | cell chemotaxis | CXCL12 | IDA | | Human | PMID:18308860 |
| Biological Process | GO:0042074 | cell migration involved in gastrulation | cxcl12a | IMP | ZFIN:ZDB-MRPHLNO-051028-2 | Zfish | PMID:18579679 |
| Biological Process | GO:0042074 | cell migration involved in gastrulation | cxcl12a | IGI | ZFIN:ZDB-MRPHLNO-051028-1,ZFIN:ZDB-MRPHLNO-051028-2 | Zfish | PMID:18579679 |
| Biological Process | GO:0042074 | cell migration involved in gastrulation | cxcl12b | IMP | ZFIN:ZDB-MRPHLNO-051028-1 | Zfish | PMID:18579679 |
| Biological Process | GO:0042074 | cell migration involved in gastrulation | cxcl12b | IGI | ZFIN:ZDB-MRPHLNO-051028-1,ZFIN:ZDB-MRPHLNO-051028-2 | Zfish | PMID:18579679 |
| Biological Process | GO:0002042 | cell migration involved in sprouting angiogenesis | cxcl12b | IGI | ZFIN:ZDB-MRPHLNO-110429-2,ZFIN:ZDB-MRPHLNO-110429-3 | Zfish | PMID:21429985 |
| Biological Process | GO:0002042 | cell migration involved in sprouting angiogenesis | cxcl12b | IMP | ZFIN:ZDB-MRPHLNO-110429-2 | Zfish | PMID:21429985 |
| Biological Process | GO:0000902 | cell morphogenesis | cxcl12a | IGI | ZFIN:ZDB-MRPHLNO-051028-1,ZFIN:ZDB-MRPHLNO-051028-2 | Zfish | PMID:18579679 |
| Biological Process | GO:0000902 | cell morphogenesis | cxcl12b | IGI | ZFIN:ZDB-MRPHLNO-051028-1,ZFIN:ZDB-MRPHLNO-051028-2 | Zfish | PMID:18579679 |
| Biological Process | GO:0098609 | cell-cell adhesion | Cxcl12 | IDA | | Mouse | PMID:20847314 |
| Biological Process | GO:1990869 | cellular response to chemokine | CXCL12 | IMP | | Human | PMID:27400149 |
| Biological Process | GO:0038146 | chemokine (C-X-C motif) ligand 12 signaling pathway | CXCL12 | IMP | | Human | PMID:27400149 |
| Biological Process | GO:0070098 | chemokine-mediated signaling pathway | CXCL12 | IDA | | Human | PMID:20388803 |
| Biological Process | GO:0006935 | chemotaxis | cxcl12a | IGI | ZFIN:ZDB-GENO-140207-11,ZFIN:ZDB-MRPHLNO-140205-5,ZFIN:ZDB-MRPHLNO-140205-6 | Zfish | PMID:24119842 |
| Biological Process | GO:0006935 | chemotaxis | cxcl12a | IMP | ZFIN:ZDB-GENO-101119-34 | Zfish | PMID:24119842 |
| Biological Process | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain | Cxcl12 | IMP | | Rat | PMID:27760212|RGD:12910551 |
| Biological Process | GO:0050965 | detection of temperature stimulus involved in sensory perception of pain | Cxcl12 | IMP | | Rat | PMID:27760212|RGD:12910551 |
| Biological Process | GO:0071542 | dopaminergic neuron differentiation | Cxcl12 | IDA | | Mouse | MGI:MGI:5700491|PMID:22535492 |
| Biological Process | GO:0031101 | fin regeneration | cxcl12a | IMP | | Zfish | PMID:19503807 |
| Biological Process | GO:0031101 | fin regeneration | cxcl12a | IDA | | Zfish | PMID:16586100 |
| Biological Process | GO:0030900 | forebrain development | cxcl12a | IMP | ZFIN:ZDB-MRPHLNO-050324-1 | Zfish | PMID:19595689 |
| Biological Process | GO:0007281 | germ cell development | Cxcl12 | IDA | | Mouse | PMID:12900445 |
| Biological Process | GO:0008354 | germ cell migration | Cxcl12 | IDA | | Mouse | PMID:12900445 |
| Biological Process | GO:0008354 | germ cell migration | cxcl12a | IMP | | Zfish | PMID:15068797 |
| Biological Process | GO:0008354 | germ cell migration | cxcl12a | IMP | ZFIN:ZDB-GENO-131017-4 | Zfish | PMID:23826390 |
| Biological Process | GO:0008354 | germ cell migration | cxcl12a | IMP | | Zfish | PMID:16129886 |
| Biological Process | GO:0008354 | germ cell migration | cxcl12a | IMP | | Zfish | PMID:12464177 |
| Biological Process | GO:0035701 | hematopoietic stem cell migration | cxcl12a | IGI | ZFIN:ZDB-GENE-040426-2132 | Zfish | PMID:28416284 |
| Biological Process | GO:0030097 | hemopoiesis | cxcl12b | IMP | ZFIN:ZDB-MRPHLNO-141113-3 | Zfish | PMID:25119043 |
| Biological Process | GO:0050930 | induction of positive chemotaxis | Cxcl12 | IDA | | Mouse | PMID:12900445 |
| Biological Process | GO:0050930 | induction of positive chemotaxis | cxcl12a | IMP | | Zfish | PMID:15068797 |
| Biological Process | GO:0050930 | induction of positive chemotaxis | cxcl12a | IMP | | Zfish | PMID:15081362 |
| Biological Process | GO:0050930 | induction of positive chemotaxis | cxcl12b | IGI | ZFIN:ZDB-GENE-030318-1 | Zfish | PMID:15081362 |
| Biological Process | GO:0033622 | integrin activation | CXCL12 | IDA | | Human | PMID:29301984 |
| Biological Process | GO:0070121 | Kupffer's vesicle development | cxcl12b | IGI | ZFIN:ZDB-GENE-980526-176 | Zfish | PMID:31430272 |
| Biological Process | GO:0070121 | Kupffer's vesicle development | cxcl12b | IMP | ZFIN:ZDB-GENO-200330-3 | Zfish | PMID:31430272 |
| Biological Process | GO:0001946 | lymphangiogenesis | cxcl12a | IMP | ZFIN:ZDB-MRPHLNO-071022-5 | Zfish | PMID:24903752 |
| Biological Process | GO:0097535 | lymphoid lineage cell migration into thymus | cxcl12a | IGI | ZFIN:ZDB-MRPHLNO-071113-1,ZFIN:ZDB-MRPHLNO-121001-4 | Zfish | PMID:22342843 |
| Biological Process | GO:0097535 | lymphoid lineage cell migration into thymus | cxcl12b | IGI | ZFIN:ZDB-MRPHLNO-071113-1,ZFIN:ZDB-MRPHLNO-121001-4 | Zfish | PMID:22342843 |
| Biological Process | GO:0097535 | lymphoid lineage cell migration into thymus | cxcl12b | IGI | ZFIN:ZDB-MRPHLNO-071113-1,ZFIN:ZDB-MRPHLNO-121001-6 | Zfish | PMID:22342843 |
| Biological Process | GO:0097535 | lymphoid lineage cell migration into thymus | cxcl12b | IMP | ZFIN:ZDB-MRPHLNO-071113-1 | Zfish | PMID:22342843 |
| Biological Process | GO:0008045 | motor neuron axon guidance | Cxcl12 | IMP | MGI:MGI:1934384 | Mouse | PMID:16129397 |
| Biological Process | GO:0030239 | myofibril assembly | cxcl12a | IMP | ZFIN:ZDB-MRPHLNO-071022-4 | Zfish | PMID:17517144 |
| Biological Process | GO:0008285 | negative regulation of cell population proliferation | Cxcl12 | IGI | MGI:MGI:109562 | Mouse | PMID:22956548 |
| Biological Process | GO:0008285 | negative regulation of cell population proliferation | Cxcl12 | IGI | MGI:MGI:1341264 | Mouse | PMID:22956548 |
| Biological Process | GO:0008285 | negative regulation of cell population proliferation | cxcl12a | IDA | | Zfish | PMID:16586100 |
| Biological Process | GO:2000669 | negative regulation of dendritic cell apoptotic process | CXCL12 | IDA | | Human | PMID:15059845 |
| Biological Process | GO:2000472 | negative regulation of hematopoietic stem cell migration | cxcl12a | IGI | ZFIN:ZDB-GENO-130204-7,ZFIN:ZDB-MRPHLNO-051028-2 | Zfish | PMID:21856868 |
| Biological Process | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage | CXCL12 | IDA | | Human | PMID:20388803 |
| Biological Process | GO:1903237 | negative regulation of leukocyte tethering or rolling | CXCL12 | IDA | | Human | PMID:18308860 |
| Biological Process | GO:0090024 | negative regulation of neutrophil chemotaxis | cxcl12a | IGI | ZFIN:ZDB-GENO-110209-3,ZFIN:ZDB-MRPHLNO-051028-3 | Zfish | PMID:20592249 |
| Biological Process | GO:0001764 | neuron migration | Cxcl12 | IDA | | Rat | PMID:12401342|RGD:1358280 |
| Biological Process | GO:0001764 | neuron migration | cxcl12a | IGI | ZFIN:ZDB-GENO-101119-34,ZFIN:ZDB-MRPHLNO-051028-1 | Zfish | PMID:23382464 |
| Biological Process | GO:0001764 | neuron migration | cxcl12a | IMP | ZFIN:ZDB-MRPHLNO-050324-1 | Zfish | PMID:19595689 |
| Biological Process | GO:0001764 | neuron migration | cxcl12a | IMP | ZFIN:ZDB-MRPHLNO-050324-1 | Zfish | PMID:15659553 |
| Biological Process | GO:0001764 | neuron migration | cxcl12a | IMP | | Zfish | PMID:16129396 |
| Biological Process | GO:0001764 | neuron migration | cxcl12b | IMP | ZFIN:ZDB-MRPHLNO-051028-1 | Zfish | PMID:16129396 |
| Biological Process | GO:0001764 | neuron migration | cxcl12b | IGI | ZFIN:ZDB-GENO-101119-34,ZFIN:ZDB-MRPHLNO-051028-1 | Zfish | PMID:23382464 |
| Biological Process | GO:0048842 | positive regulation of axon extension involved in axon guidance | Cxcl12 | IMP | | Rat | PMID:18701697|RGD:2306571 |
| Biological Process | GO:0048842 | positive regulation of axon extension involved in axon guidance | cxcl12a | IDA | | Zfish | PMID:17267551 |
| Biological Process | GO:0090280 | positive regulation of calcium ion import | Cxcl12 | IDA | | Mouse | PMID:8962122 |
| Biological Process | GO:0045785 | positive regulation of cell adhesion | CXCL12 | IDA | | Human | PMID:23620790 |
| Biological Process | GO:0030335 | positive regulation of cell migration | Cxcl12 | IDA | | Mouse | PMID:12900445 |
| Biological Process | GO:0030335 | positive regulation of cell migration | Cxcl12 | IMP | | Rat | PMID:11820456|RGD:632503 |
| Biological Process | GO:0030335 | positive regulation of cell migration | Cxcl12 | IMP | | Rat | PMID:19327017|RGD:2306564 |
| Biological Process | GO:0030335 | positive regulation of cell migration | cxcl12a | IMP | ZFIN:ZDB-GENO-070123-6 | Zfish | PMID:16678780 |
| Biological Process | GO:0030335 | positive regulation of cell migration | cxcl12a | IMP | ZFIN:ZDB-MRPHLNO-050324-1 | Zfish | PMID:17267445 |
| Biological Process | GO:0030335 | positive regulation of cell migration | cxcl12a | IMP | | Zfish | PMID:12444253 |
| Biological Process | GO:0008284 | positive regulation of cell population proliferation | Cxcl12 | IMP | | Rat | PMID:17425785|RGD:2306580 |
| Biological Process | GO:0050921 | positive regulation of chemotaxis | Cxcl12 | IGI | MGI:MGI:1341264 | Mouse | PMID:22956548 |
| Biological Process | GO:0033603 | positive regulation of dopamine secretion | Cxcl12 | IDA | | Rat | PMID:18671738|RGD:2306573 |
| Biological Process | GO:0001938 | positive regulation of endothelial cell proliferation | Cxcl12 | IMP | | Rat | PMID:18690419|RGD:2306572 |
| Biological Process | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | Cxcl12 | IGI | MGI:MGI:1341264 | Mouse | PMID:22956548 |
| Biological Process | GO:0090026 | positive regulation of monocyte chemotaxis | CXCL12 | IDA | | Human | PMID:18802065 |
| Biological Process | GO:1901741 | positive regulation of myoblast fusion | Cxcl12 | IMP | | Mouse | PMID:23844157 |
| Biological Process | GO:0045666 | positive regulation of neuron differentiation | Cxcl12 | IMP | | Rat | PMID:17425785|RGD:2306580 |
| Biological Process | GO:0051897 | positive regulation of protein kinase B signaling | Cxcl12 | IGI | MGI:MGI:1341264 | Mouse | PMID:22956548 |
| Biological Process | GO:2000406 | positive regulation of T cell migration | CXCL12 | IDA | | Human | PMID:23620790 |
| Biological Process | GO:0048920 | posterior lateral line neuromast primordium migration | cxcl12a | IMP | ZFIN:ZDB-GENO-101119-34 | Zfish | PMID:24119842 |
| Biological Process | GO:0048920 | posterior lateral line neuromast primordium migration | cxcl12a | IMP | ZFIN:ZDB-MRPHLNO-051028-3 | Zfish | PMID:29945870 |
| Biological Process | GO:0048920 | posterior lateral line neuromast primordium migration | cxcl12a | IMP | ZFIN:ZDB-MRPHLNO-050324-1 | Zfish | PMID:17394634 |
| Biological Process | GO:0048920 | posterior lateral line neuromast primordium migration | cxcl12a | IGI | ZFIN:ZDB-MRPHLNO-050324-1,ZFIN:ZDB-MRPHLNO-081216-1 | Zfish | PMID:18585376 |
| Biological Process | GO:0048920 | posterior lateral line neuromast primordium migration | cxcl12a | IGI | ZFIN:ZDB-GENO-140207-11,ZFIN:ZDB-MRPHLNO-140205-5,ZFIN:ZDB-MRPHLNO-140205-6 | Zfish | PMID:24119842 |
| Biological Process | GO:0048920 | posterior lateral line neuromast primordium migration | cxcl12a | IGI | ZFIN:ZDB-MRPHLNO-050324-1,ZFIN:ZDB-MRPHLNO-100608-9 | Zfish | PMID:20308561 |
| Biological Process | GO:0048920 | posterior lateral line neuromast primordium migration | cxcl12a | IMP | ZFIN:ZDB-GENO-101119-34 | Zfish | PMID:17570670 |
| Biological Process | GO:0048920 | posterior lateral line neuromast primordium migration | cxcl12a | IMP | ZFIN:ZDB-MRPHLNO-050324-1 | Zfish | PMID:20308561 |
| Biological Process | GO:0030516 | regulation of axon extension | cxcl12a | IMP | | Zfish | PMID:15081362 |
| Biological Process | GO:0051924 | regulation of calcium ion transport | Cxcl12 | IDA | | Rat | PMID:18671738|RGD:2306573 |
| Biological Process | GO:0030334 | regulation of cell migration | Cxcl12 | IGI | MGI:MGI:1306805 | Mouse | PMID:19064997 |
| Biological Process | GO:0030334 | regulation of cell migration | Cxcl12 | IDA | | Mouse | PMID:12183377 |
| Biological Process | GO:0106014 | regulation of inflammatory response to wounding | cxcl12a | IMP | ZFIN:ZDB-GENO-101119-34 | Zfish | PMID:28220184 |
| Biological Process | GO:0051453 | regulation of intracellular pH | cxcl12a | IGI | ZFIN:ZDB-GENE-030721-1 | Zfish | PMID:25843033 |
| Biological Process | GO:0051453 | regulation of intracellular pH | cxcl12b | IGI | ZFIN:ZDB-GENE-030318-1 | Zfish | PMID:25843033 |
| Biological Process | GO:0090022 | regulation of neutrophil chemotaxis | cxcl12a | IMP | ZFIN:ZDB-GENO-101119-34 | Zfish | PMID:28220184 |
| Biological Process | GO:0009408 | response to heat | Cxcl12 | IEP | | Rat | PMID:18552407|RGD:2306574 |
| Biological Process | GO:0001666 | response to hypoxia | Cxcl12 | IEP | | Rat | PMID:17581219|RGD:2306578 |
| Biological Process | GO:0009612 | response to mechanical stimulus | Cxcl12 | IEP | | Rat | PMID:17145991|RGD:2306582 |
| Biological Process | GO:0043434 | response to peptide hormone | Cxcl12 | IEP | | Rat | PMID:19294511|RGD:2306566 |
| Biological Process | GO:0009314 | response to radiation | Cxcl12 | IEP | | Rat | PMID:19024651|RGD:2306569 |
| Biological Process | GO:1990478 | response to ultrasound | Cxcl12 | IEP | | Rat | PMID:25181476|RGD:11352664 |
| Biological Process | GO:0031290 | retinal ganglion cell axon guidance | cxcl12a | IDA | | Zfish | PMID:17267551 |
| Biological Process | GO:0031290 | retinal ganglion cell axon guidance | cxcl12a | IGI | ZFIN:ZDB-GENO-070412-4,ZFIN:ZDB-MRPHLNO-050318-6 | Zfish | PMID:17267551 |
| Biological Process | GO:0031290 | retinal ganglion cell axon guidance | cxcl12b | IGI | ZFIN:ZDB-GENO-070412-4,ZFIN:ZDB-MRPHLNO-051028-1 | Zfish | PMID:17267551 |
| Biological Process | GO:0002040 | sprouting angiogenesis | cxcl12b | IMP | ZFIN:ZDB-GENO-110513-4 | Zfish | PMID:21429983 |
| Biological Process | GO:0055002 | striated muscle cell development | cxcl12a | IMP | ZFIN:ZDB-MRPHLNO-071022-4 | Zfish | PMID:17517144 |
| Biological Process | GO:0042098 | T cell proliferation | Cxcl12 | IMP | MGI:MGI:1934384 | Mouse | PMID:12707343 |
| Biological Process | GO:0022029 | telencephalon cell migration | Cxcl12 | IMP | | Rat | PMID:16971524|RGD:2306584 |
| Biological Process | GO:0021730 | trigeminal sensory nucleus development | cxcl12a | IGI | ZFIN:ZDB-GENO-101119-34,ZFIN:ZDB-MRPHLNO-051028-1 | Zfish | PMID:23382464 |
| Biological Process | GO:0021730 | trigeminal sensory nucleus development | cxcl12b | IGI | ZFIN:ZDB-GENO-101119-34,ZFIN:ZDB-MRPHLNO-051028-1 | Zfish | PMID:23382464 |
| Biological Process | GO:0001570 | vasculogenesis | cxcl12b | IMP | ZFIN:ZDB-MRPHLNO-051028-1 | Zfish | PMID:19797767 |
 Analysis Tools
Analysis Tools