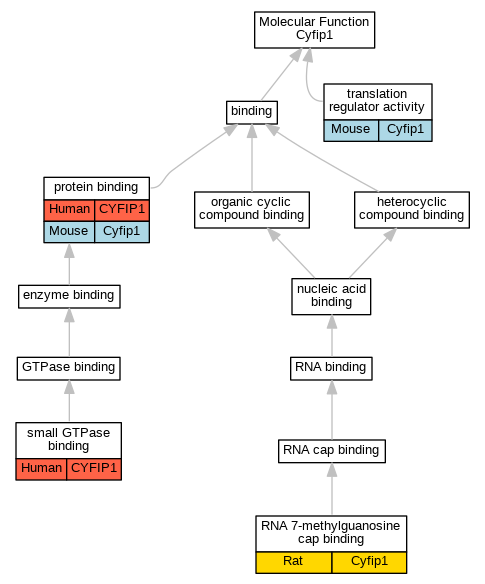
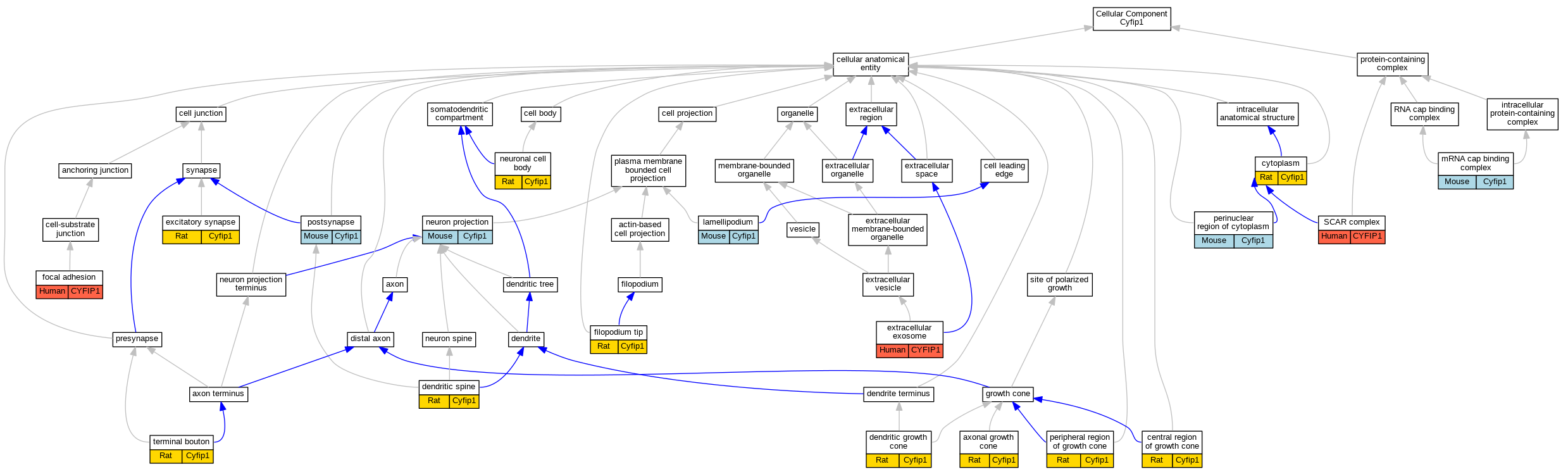
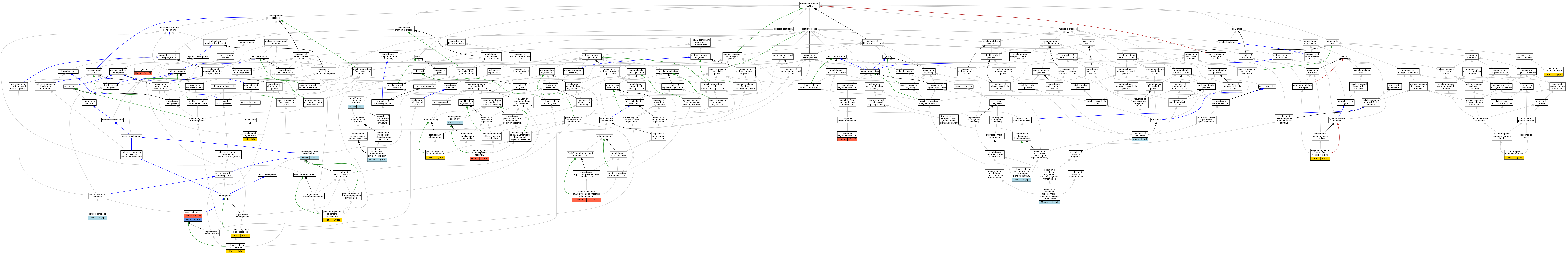
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0005515 | protein binding | CYFIP1 | IPI | UniProtKB:Q06787 | Human | PMID:11438699 |
| Molecular Function | GO:0005515 | protein binding | CYFIP1 | IPI | UniProtKB:Q9Y2A7 | Human | PMID:15294869 |
| Molecular Function | GO:0005515 | protein binding | CYFIP1 | IPI | UniProtKB:Q9Y2A7 | Human | PMID:15296760 |
| Molecular Function | GO:0005515 | protein binding | CYFIP1 | IPI | UniProtKB:Q16555 | Human | PMID:16260607 |
| Molecular Function | GO:0005515 | protein binding | CYFIP1 | IPI | UniProtKB:P63073 | Human | PMID:18805096 |
| Molecular Function | GO:0005515 | protein binding | CYFIP1 | IPI | UniProtKB:Q9Y2A7 | Human | PMID:19363480 |
| Molecular Function | GO:0005515 | protein binding | CYFIP1 | IPI | UniProtKB:Q9Y2A7 | Human | PMID:21107423 |
| Molecular Function | GO:0005515 | protein binding | CYFIP1 | IPI | UniProtKB:B1Q236,UniProtKB:Q8WUW1,UniProtKB:Q92558,UniProtKB:Q9NYB9,UniProtKB:Q9Y2A7 | Human | PMID:24439377 |
| Molecular Function | GO:0005515 | protein binding | CYFIP1 | IPI | UniProtKB:O14901 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | CYFIP1 | IPI | UniProtKB:O60333-2 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | CYFIP1 | IPI | UniProtKB:O76024 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | CYFIP1 | IPI | UniProtKB:P01023 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | CYFIP1 | IPI | UniProtKB:P07196 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | CYFIP1 | IPI | UniProtKB:P51608 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | CYFIP1 | IPI | UniProtKB:Q7Z699 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | Cyfip1 | IPI | UniProtKB:P35922 | Mouse | MGI:MGI:3036554|PMID:11438699 |
| Molecular Function | GO:0005515 | protein binding | Cyfip1 | IPI | UniProtKB:B1Q236,UniProtKB:Q8R5H6 | Mouse | MGI:MGI:5688823|PMID:24439377 |
| Molecular Function | GO:0005515 | protein binding | Cyfip1 | IPI | UniProtKB:P35922|UniProtKB:P63073 | Mouse | MGI:MGI:4418608|PMID:18805096 |
| Molecular Function | GO:0005515 | protein binding | Cyfip1 | IPI | UniProtKB:Q8BHE4 | Mouse | MGI:MGI:5825407|PMID:27605705 |
| Molecular Function | GO:0005515 | protein binding | Cyfip1 | IPI | UniProtKB:P63073 | Mouse | MGI:MGI:4418608|PMID:18805096 |
| Molecular Function | GO:0005515 | protein binding | Cyfip1 | IPI | UniProtKB:Q5DU14|UniProtKB:Q6PFX7|UniProtKB:Q8BM65 | Mouse | MGI:MGI:5304984|PMID:21946561 |
| Molecular Function | GO:0000340 | RNA 7-methylguanosine cap binding | Cyfip1 | IDA | | Rat | PMID:25453757|RGD:11558007 |
| Molecular Function | GO:0031267 | small GTPase binding | CYFIP1 | IMP | | Human | PMID:21107423 |
| Molecular Function | GO:0045182 | translation regulator activity | Cyfip1 | EXP | | Mouse | PMID:24050404 |
| Molecular Function | GO:0045182 | translation regulator activity | Cyfip1 | IDA | | Mouse | PMID:24050404 |
| Molecular Function | GO:0045182 | translation regulator activity | Cyfip1 | IPI | | Mouse | PMID:24050404 |
| Molecular Function | GO:0045182 | translation regulator activity | Cyfip1 | IMP | | Mouse | PMID:24050404 |
| Molecular Function | GO:0045182 | translation regulator activity | Cyfip1 | IEP | | Mouse | PMID:24050404 |
| Cellular Component | GO:0044295 | axonal growth cone | Cyfip1 | IDA | | Rat | PMID:16260607|RGD:11557987 |
| Cellular Component | GO:0090724 | central region of growth cone | Cyfip1 | IDA | | Rat | PMID:16260607|RGD:11557987 |
| Cellular Component | GO:0005737 | cytoplasm | Cyfip1 | IDA | | Rat | PMID:26000921|RGD:11568065 |
| Cellular Component | GO:0044294 | dendritic growth cone | Cyfip1 | IDA | | Rat | PMID:16260607|RGD:11557987 |
| Cellular Component | GO:0043197 | dendritic spine | Cyfip1 | IDA | | Rat | PMID:24667445|RGD:11568064 |
| Cellular Component | GO:0060076 | excitatory synapse | Cyfip1 | IDA | | Rat | PMID:24667445|RGD:11568064 |
| Cellular Component | GO:0070062 | extracellular exosome | CYFIP1 | HDA | | Human | PMID:20458337 |
| Cellular Component | GO:0070062 | extracellular exosome | CYFIP1 | HDA | | Human | PMID:23533145 |
| Cellular Component | GO:0032433 | filopodium tip | Cyfip1 | IDA | | Rat | PMID:16260607|RGD:11557987 |
| Cellular Component | GO:0005925 | focal adhesion | CYFIP1 | HDA | | Human | PMID:21423176 |
| Cellular Component | GO:0030027 | lamellipodium | Cyfip1 | IDA | | Mouse | PMID:14765121 |
| Cellular Component | GO:0005845 | mRNA cap binding complex | Cyfip1 | IDA | | Mouse | MGI:MGI:4418608|PMID:18805096 |
| Cellular Component | GO:0043005 | neuron projection | Cyfip1 | IDA | | Mouse | MGI:MGI:3036554|PMID:11438699 |
| Cellular Component | GO:0043025 | neuronal cell body | Cyfip1 | IDA | | Rat | PMID:16260607|RGD:11557987 |
| Cellular Component | GO:0048471 | perinuclear region of cytoplasm | Cyfip1 | IDA | | Mouse | MGI:MGI:3036554|PMID:11438699 |
| Cellular Component | GO:0090725 | peripheral region of growth cone | Cyfip1 | IDA | | Rat | PMID:16260607|RGD:11557987 |
| Cellular Component | GO:0098794 | postsynapse | Cyfip1 | IDA | | Mouse | PMID:24050404 |
| Cellular Component | GO:0031209 | SCAR complex | CYFIP1 | IMP | | Human | PMID:21107423 |
| Cellular Component | GO:0043195 | terminal bouton | Cyfip1 | IDA | | Rat | PMID:26843638|RGD:11342679 |
| Biological Process | GO:0048675 | axon extension | CYFIP1 | IMP | | Human | PMID:16260607 |
| Biological Process | GO:0048675 | axon extension | cyfip1 | IMP | ZFIN:ZDB-CRISPR-191120-4 | Zfish | PMID:29518358 |
| Biological Process | GO:0032869 | cellular response to insulin stimulus | Cyfip1 | IMP | | Rat | PMID:16260607|RGD:11557987 |
| Biological Process | GO:0050890 | cognition | CYFIP1 | IMP | | Human | PMID:17519220 |
| Biological Process | GO:0097484 | dendrite extension | Cyfip1 | IMP | | Mouse | MGI:MGI:5825407|PMID:27605705 |
| Biological Process | GO:0030032 | lamellipodium assembly | Cyfip1 | IDA | | Mouse | PMID:14765121 |
| Biological Process | GO:0099563 | modification of synaptic structure | Cyfip1 | IMP | | Mouse | MGI:MGI:5515833|PMID:24050404 |
| Biological Process | GO:1903422 | negative regulation of synaptic vesicle recycling | Cyfip1 | IMP | | Rat | PMID:26843638|RGD:11342679 |
| Biological Process | GO:0031175 | neuron projection development | Cyfip1 | IMP | | Mouse | MGI:MGI:5825407|PMID:27605705 |
| Biological Process | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation | CYFIP1 | IMP | | Human | PMID:21107423 |
| Biological Process | GO:0045773 | positive regulation of axon extension | Cyfip1 | IMP | | Rat | PMID:16260607|RGD:11557987 |
| Biological Process | GO:0050772 | positive regulation of axonogenesis | Cyfip1 | IMP | | Rat | PMID:16260607|RGD:11557987 |
| Biological Process | GO:1900006 | positive regulation of dendrite development | Cyfip1 | IMP | | Rat | PMID:16260607|RGD:11557987 |
| Biological Process | GO:0010592 | positive regulation of lamellipodium assembly | CYFIP1 | IDA | | Human | PMID:21107423 |
| Biological Process | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway | Cyfip1 | IMP | | Mouse | MGI:MGI:5825407|PMID:27605705 |
| Biological Process | GO:1900029 | positive regulation of ruffle assembly | Cyfip1 | IMP | | Rat | PMID:16260607|RGD:11557987 |
| Biological Process | GO:0016601 | Rac protein signal transduction | CYFIP1 | IMP | | Human | PMID:21107423 |
| Biological Process | GO:1905274 | regulation of modification of postsynaptic actin cytoskeleton | Cyfip1 | IMP | | Mouse | PMID:24050404 |
| Biological Process | GO:1905274 | regulation of modification of postsynaptic actin cytoskeleton | Cyfip1 | IDA | | Mouse | PMID:24050404 |
| Biological Process | GO:0031641 | regulation of myelination | Cyfip1 | IMP | | Rat | PMID:31371763|RGD:14981598 |
| Biological Process | GO:0006417 | regulation of translation | Cyfip1 | IMP | | Mouse | MGI:MGI:5515833|PMID:24050404 |
| Biological Process | GO:0099578 | regulation of translation at postsynapse, modulating synaptic transmission | Cyfip1 | IPI | | Mouse | PMID:24050404 |
| Biological Process | GO:0099578 | regulation of translation at postsynapse, modulating synaptic transmission | Cyfip1 | EXP | | Mouse | PMID:24050404 |
| Biological Process | GO:0099578 | regulation of translation at postsynapse, modulating synaptic transmission | Cyfip1 | IDA | | Mouse | PMID:24050404 |
| Biological Process | GO:0099578 | regulation of translation at postsynapse, modulating synaptic transmission | Cyfip1 | IMP | | Mouse | PMID:24050404 |
| Biological Process | GO:0099578 | regulation of translation at postsynapse, modulating synaptic transmission | Cyfip1 | IEP | | Mouse | PMID:24050404 |
| Biological Process | GO:0051602 | response to electrical stimulus | Cyfip1 | IDA | | Rat | PMID:25453757|RGD:11558007 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
 Analysis Tools
Analysis Tools