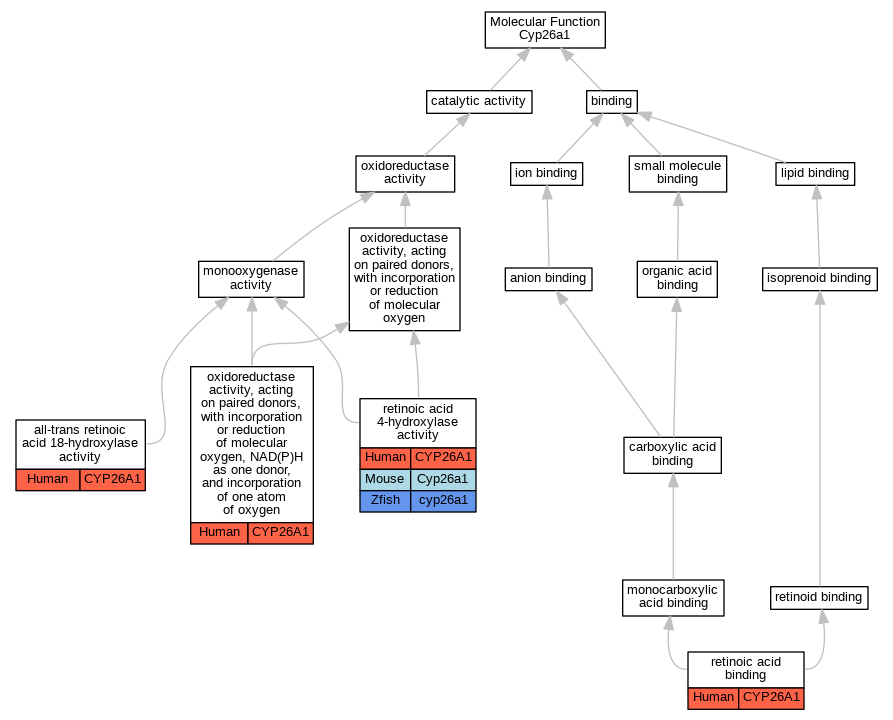
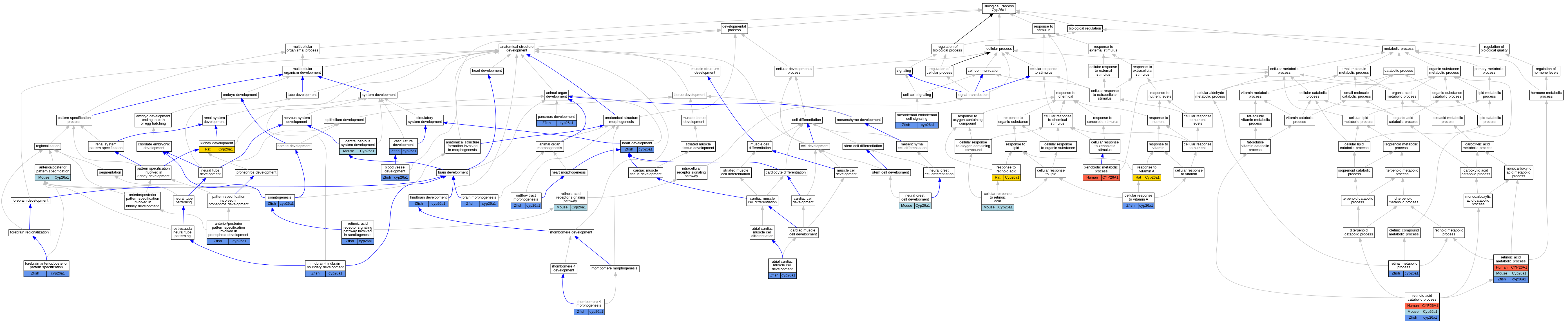
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0062183 | all-trans retinoic acid 18-hydroxylase activity | CYP26A1 | IDA | | Human | PMID:22020119 |
| Molecular Function | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen | CYP26A1 | IDA | | Human | PMID:26937021 |
| Molecular Function | GO:0008401 | retinoic acid 4-hydroxylase activity | CYP26A1 | IDA | | Human | PMID:10823918 |
| Molecular Function | GO:0008401 | retinoic acid 4-hydroxylase activity | CYP26A1 | IDA | | Human | PMID:9228017 |
| Molecular Function | GO:0008401 | retinoic acid 4-hydroxylase activity | CYP26A1 | IDA | | Human | PMID:9716180 |
| Molecular Function | GO:0008401 | retinoic acid 4-hydroxylase activity | Cyp26a1 | IDA | | Mouse | MGI:MGI:1195386|PMID:9442090 |
| Molecular Function | GO:0008401 | retinoic acid 4-hydroxylase activity | Cyp26a1 | IDA | | Mouse | PMID:9250660 |
| Molecular Function | GO:0008401 | retinoic acid 4-hydroxylase activity | cyp26a1 | IDA | | Zfish | PMID:8939936 |
| Molecular Function | GO:0001972 | retinoic acid binding | CYP26A1 | IDA | | Human | PMID:10823918 |
| Biological Process | GO:0009952 | anterior/posterior pattern specification | Cyp26a1 | IGI | MGI:MGI:2679699 | Mouse | PMID:17067568 |
| Biological Process | GO:0009952 | anterior/posterior pattern specification | Cyp26a1 | IMP | MGI:MGI:2183292 | Mouse | PMID:17067568 |
| Biological Process | GO:0034672 | anterior/posterior pattern specification involved in pronephros development | cyp26a1 | IMP | ZFIN:ZDB-MRPHLNO-050316-1 | Zfish | PMID:27406002 |
| Biological Process | GO:0055014 | atrial cardiac muscle cell development | cyp26a1 | IGI | ZFIN:ZDB-MRPHLNO-070410-6,ZFIN:ZDB-MRPHLNO-131002-1,ZFIN:ZDB-MRPHLNO-131002-2 | Zfish | PMID:24667328 |
| Biological Process | GO:0055014 | atrial cardiac muscle cell development | cyp26a1 | IGI | ZFIN:ZDB-GENO-070411-2,ZFIN:ZDB-MRPHLNO-070410-6 | Zfish | PMID:24667328 |
| Biological Process | GO:0001568 | blood vessel development | cyp26a1 | IMP | ZFIN:ZDB-GENO-050324-1 | Zfish | PMID:15680360 |
| Biological Process | GO:0001568 | blood vessel development | cyp26a1 | IMP | ZFIN:ZDB-MRPHLNO-050316-1 | Zfish | PMID:15680360 |
| Biological Process | GO:0048854 | brain morphogenesis | cyp26a1 | IMP | ZFIN:ZDB-GENO-070411-2 | Zfish | PMID:17998248 |
| Biological Process | GO:0071300 | cellular response to retinoic acid | Cyp26a1 | IEP | | Mouse | MGI:MGI:3809313|PMID:18816858 |
| Biological Process | GO:0071299 | cellular response to vitamin A | cyp26a1 | IDA | | Zfish | PMID:17253779 |
| Biological Process | GO:0007417 | central nervous system development | Cyp26a1 | IMP | MGI:MGI:2183292 | Mouse | PMID:17067568 |
| Biological Process | GO:0007417 | central nervous system development | Cyp26a1 | IGI | MGI:MGI:2679699 | Mouse | PMID:17067568 |
| Biological Process | GO:0021797 | forebrain anterior/posterior pattern specification | cyp26a1 | IGI | ZFIN:ZDB-GENE-990415-44,ZFIN:ZDB-MRPHLNO-080501-1 | Zfish | PMID:17998248 |
| Biological Process | GO:0007507 | heart development | cyp26a1 | IMP | ZFIN:ZDB-MRPHLNO-131002-1,ZFIN:ZDB-MRPHLNO-131002-2 | Zfish | PMID:23990796 |
| Biological Process | GO:0007507 | heart development | cyp26a1 | IGI | ZFIN:ZDB-MRPHLNO-131002-1,ZFIN:ZDB-MRPHLNO-131002-2,ZFIN:ZDB-MRPHLNO-131018-1 | Zfish | PMID:23990796 |
| Biological Process | GO:0030902 | hindbrain development | cyp26a1 | IMP | ZFIN:ZDB-GENO-050324-1 | Zfish | PMID:15680360 |
| Biological Process | GO:0030902 | hindbrain development | cyp26a1 | IGI | ZFIN:ZDB-GENO-070411-2,ZFIN:ZDB-MRPHLNO-070410-1,ZFIN:ZDB-MRPHLNO-070410-6 | Zfish | PMID:17164423 |
| Biological Process | GO:0001822 | kidney development | Cyp26a1 | IEP | | Rat | PMID:15567713|RGD:2306322 |
| Biological Process | GO:0003131 | mesodermal-endodermal cell signaling | cyp26a1 | IMP | ZFIN:ZDB-GENO-070411-2 | Zfish | PMID:19416885 |
| Biological Process | GO:0030917 | midbrain-hindbrain boundary development | cyp26a1 | IGI | ZFIN:ZDB-MRPHLNO-131002-1,ZFIN:ZDB-MRPHLNO-131002-2,ZFIN:ZDB-MRPHLNO-131018-1 | Zfish | PMID:23990796 |
| Biological Process | GO:0014032 | neural crest cell development | Cyp26a1 | IGI | MGI:MGI:2679699 | Mouse | PMID:17067568 |
| Biological Process | GO:0003151 | outflow tract morphogenesis | cyp26a1 | IGI | ZFIN:ZDB-MRPHLNO-070410-6,ZFIN:ZDB-MRPHLNO-131002-1 | Zfish | PMID:27893754 |
| Biological Process | GO:0031016 | pancreas development | cyp26a1 | IGI | ZFIN:ZDB-GENO-070411-2,ZFIN:ZDB-MRPHLNO-070410-1,ZFIN:ZDB-MRPHLNO-070410-6 | Zfish | PMID:19416885 |
| Biological Process | GO:0031016 | pancreas development | cyp26a1 | IMP | ZFIN:ZDB-GENO-070411-2 | Zfish | PMID:19416885 |
| Biological Process | GO:0032526 | response to retinoic acid | Cyp26a1 | IEP | | Rat | PMID:19700416|RGD:6484694 |
| Biological Process | GO:0033189 | response to vitamin A | Cyp26a1 | IEP | | Rat | PMID:19700416|RGD:6484694 |
| Biological Process | GO:0042574 | retinal metabolic process | cyp26a1 | IMP | ZFIN:ZDB-GENO-070411-2 | Zfish | PMID:17164423 |
| Biological Process | GO:0034653 | retinoic acid catabolic process | CYP26A1 | IDA | | Human | PMID:10823918 |
| Biological Process | GO:0034653 | retinoic acid catabolic process | Cyp26a1 | IDA | | Mouse | PMID:9250660 |
| Biological Process | GO:0034653 | retinoic acid catabolic process | cyp26a1 | IMP | | Zfish | PMID:19416885 |
| Biological Process | GO:0042573 | retinoic acid metabolic process | CYP26A1 | IDA | | Human | PMID:22020119 |
| Biological Process | GO:0042573 | retinoic acid metabolic process | CYP26A1 | IDA | | Human | PMID:9716180 |
| Biological Process | GO:0042573 | retinoic acid metabolic process | Cyp26a1 | IDA | | Mouse | MGI:MGI:1195386|PMID:9442090 |
| Biological Process | GO:0042573 | retinoic acid metabolic process | Cyp26a1 | IDA | | Mouse | MGI:MGI:894533|PMID:9250660 |
| Biological Process | GO:0042573 | retinoic acid metabolic process | Cyp26a1 | IMP | MGI:MGI:2183292 | Mouse | PMID:15531370 |
| Biological Process | GO:0042573 | retinoic acid metabolic process | cyp26a1 | IDA | | Zfish | PMID:8939936 |
| Biological Process | GO:0042573 | retinoic acid metabolic process | cyp26a1 | IDA | | Zfish | PMID:16455818 |
| Biological Process | GO:0042573 | retinoic acid metabolic process | cyp26a1 | IMP | ZFIN:ZDB-GENO-070411-2 | Zfish | PMID:17164423 |
| Biological Process | GO:0048384 | retinoic acid receptor signaling pathway | Cyp26a1 | IDA | | Mouse | PMID:9250660 |
| Biological Process | GO:0048384 | retinoic acid receptor signaling pathway | Cyp26a1 | IGI | MGI:MGI:2679699 | Mouse | PMID:17067568 |
| Biological Process | GO:0090242 | retinoic acid receptor signaling pathway involved in somitogenesis | cyp26a1 | IMP | ZFIN:ZDB-MRPHLNO-140205-4 | Zfish | PMID:23975936 |
| Biological Process | GO:0021661 | rhombomere 4 morphogenesis | cyp26a1 | IGI | ZFIN:ZDB-GENO-070411-2,ZFIN:ZDB-MRPHLNO-070410-1 | Zfish | PMID:17164423 |
| Biological Process | GO:0021661 | rhombomere 4 morphogenesis | cyp26a1 | IGI | ZFIN:ZDB-GENO-070411-2,ZFIN:ZDB-MRPHLNO-070410-6 | Zfish | PMID:17164423 |
| Biological Process | GO:0021661 | rhombomere 4 morphogenesis | cyp26a1 | IGI | ZFIN:ZDB-GENO-070411-2,ZFIN:ZDB-MRPHLNO-070410-1,ZFIN:ZDB-MRPHLNO-070410-6 | Zfish | PMID:17164423 |
| Biological Process | GO:0021661 | rhombomere 4 morphogenesis | cyp26a1 | IMP | ZFIN:ZDB-GENO-070411-2 | Zfish | PMID:17164423 |
| Biological Process | GO:0001756 | somitogenesis | cyp26a1 | IMP | ZFIN:ZDB-MRPHLNO-050316-1 | Zfish | PMID:17098223 |
| Biological Process | GO:0001944 | vasculature development | cyp26a1 | IGI | ZFIN:ZDB-MRPHLNO-070410-6,ZFIN:ZDB-MRPHLNO-131002-1,ZFIN:ZDB-MRPHLNO-131002-2 | Zfish | PMID:24667328 |
| Biological Process | GO:0001944 | vasculature development | cyp26a1 | IGI | ZFIN:ZDB-GENO-070411-2,ZFIN:ZDB-MRPHLNO-070410-6 | Zfish | PMID:24667328 |
| Biological Process | GO:0006805 | xenobiotic metabolic process | CYP26A1 | IDA | | Human | PMID:26937021 |
 Analysis Tools
Analysis Tools