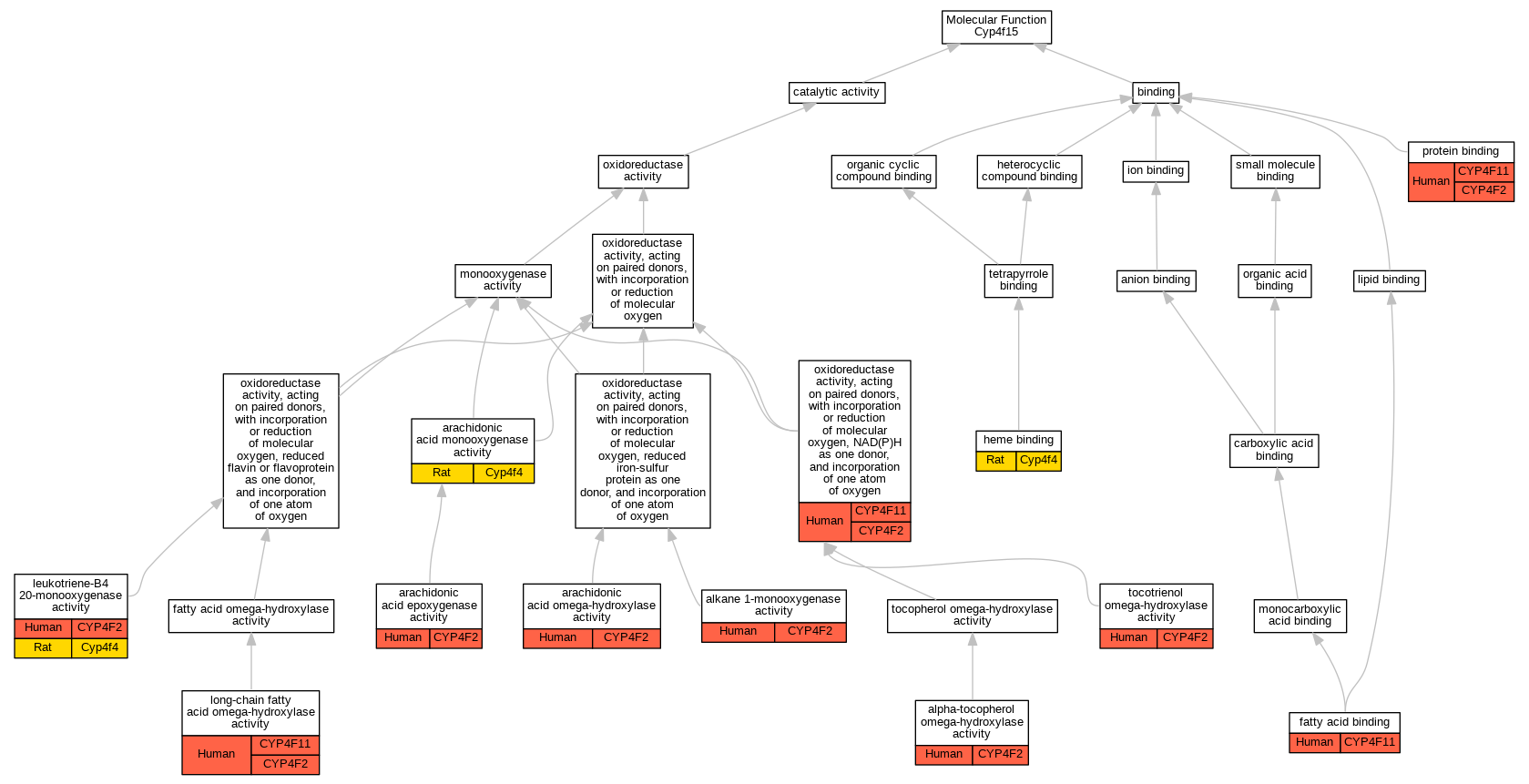
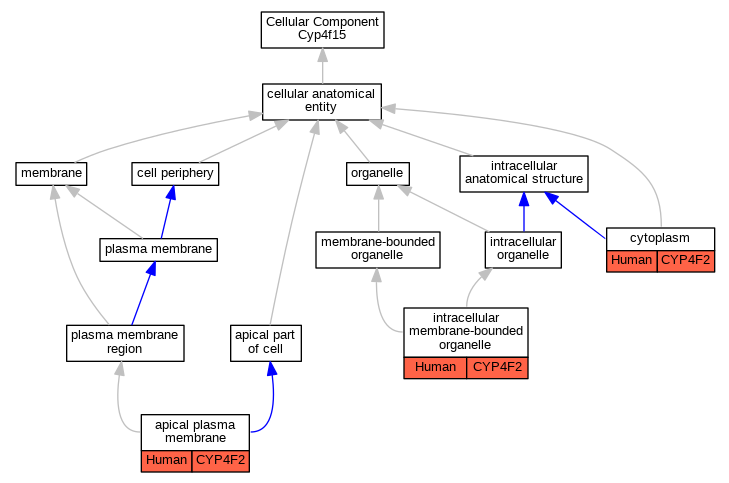
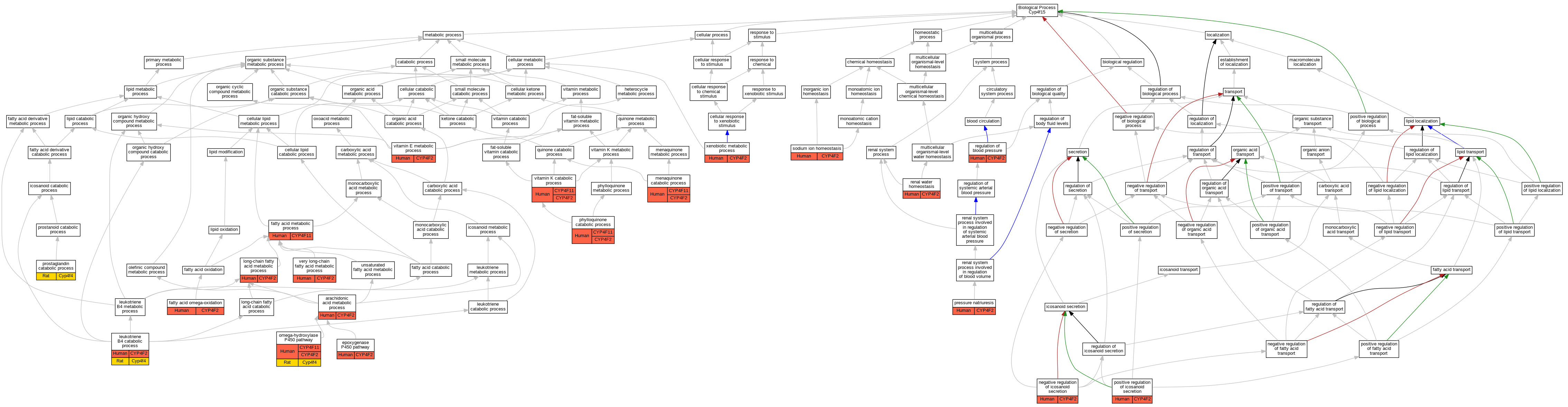
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0018685 | alkane 1-monooxygenase activity | CYP4F2 | IDA | | Human | PMID:18433732 |
| Molecular Function | GO:0052871 | alpha-tocopherol omega-hydroxylase activity | CYP4F2 | IDA | | Human | PMID:20861217 |
| Molecular Function | GO:0008392 | arachidonic acid epoxygenase activity | CYP4F2 | IDA | | Human | PMID:20861217 |
| Molecular Function | GO:0008392 | arachidonic acid epoxygenase activity | CYP4F2 | IDA | | Human | PMID:9618440 |
| Molecular Function | GO:0008391 | arachidonic acid monooxygenase activity | Cyp4f4 | IDA | | Rat | PMID:14634044|RGD:2301710 |
| Molecular Function | GO:0052869 | arachidonic acid omega-hydroxylase activity | CYP4F2 | IDA | | Human | PMID:10660572 |
| Molecular Function | GO:0052869 | arachidonic acid omega-hydroxylase activity | CYP4F2 | IDA | | Human | PMID:9618440 |
| Molecular Function | GO:0005504 | fatty acid binding | CYP4F11 | IDA | | Human | PMID:18065749 |
| Molecular Function | GO:0020037 | heme binding | Cyp4f4 | IDA | | Rat | PMID:11980497|RGD:2301711 |
| Molecular Function | GO:0050051 | leukotriene-B4 20-monooxygenase activity | CYP4F2 | IDA | | Human | PMID:8026587 |
| Molecular Function | GO:0050051 | leukotriene-B4 20-monooxygenase activity | CYP4F2 | IDA | | Human | PMID:9799565 |
| Molecular Function | GO:0050051 | leukotriene-B4 20-monooxygenase activity | Cyp4f4 | IDA | | Rat | PMID:14634044|RGD:2301710 |
| Molecular Function | GO:0050051 | leukotriene-B4 20-monooxygenase activity | Cyp4f4 | IDA | | Rat | PMID:9344476|RGD:15090848 |
| Molecular Function | GO:0102033 | long-chain fatty acid omega-hydroxylase activity | CYP4F2 | IDA | | Human | PMID:15145985 |
| Molecular Function | GO:0102033 | long-chain fatty acid omega-hydroxylase activity | CYP4F2 | IDA | | Human | PMID:18577768 |
| Molecular Function | GO:0102033 | long-chain fatty acid omega-hydroxylase activity | CYP4F11 | IDA | | Human | PMID:15364545 |
| Molecular Function | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen | CYP4F2 | IDA | | Human | PMID:19297519 |
| Molecular Function | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen | CYP4F2 | IDA | | Human | PMID:24138531 |
| Molecular Function | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen | CYP4F11 | IDA | | Human | PMID:18065749 |
| Molecular Function | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen | CYP4F11 | IDA | | Human | PMID:24138531 |
| Molecular Function | GO:0005515 | protein binding | CYP4F2 | IPI | UniProtKB:P16333 | Human | PMID:17474147 |
| Molecular Function | GO:0005515 | protein binding | CYP4F2 | IPI | UniProtKB:O43889-2 | Human | PMID:25910212 |
| Molecular Function | GO:0005515 | protein binding | CYP4F2 | IPI | UniProtKB:O15173 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CYP4F2 | IPI | UniProtKB:O95214 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CYP4F2 | IPI | UniProtKB:P15941-11 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CYP4F2 | IPI | UniProtKB:P48165 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CYP4F2 | IPI | UniProtKB:Q03395 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CYP4F2 | IPI | UniProtKB:Q12999 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CYP4F2 | IPI | UniProtKB:Q13520 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CYP4F2 | IPI | UniProtKB:Q15125 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CYP4F2 | IPI | UniProtKB:Q15546 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CYP4F2 | IPI | UniProtKB:Q3KNW5 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CYP4F2 | IPI | UniProtKB:Q53GL0 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CYP4F2 | IPI | UniProtKB:Q7KZS0 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CYP4F2 | IPI | UniProtKB:Q7Z5P4 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CYP4F2 | IPI | UniProtKB:Q7Z7G2 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CYP4F2 | IPI | UniProtKB:Q86VR2 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CYP4F2 | IPI | UniProtKB:Q8N5M9 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CYP4F2 | IPI | UniProtKB:Q8NDX2-2 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CYP4F2 | IPI | UniProtKB:Q8WY91 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CYP4F2 | IPI | UniProtKB:Q96BA8 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CYP4F2 | IPI | UniProtKB:Q96DZ7 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CYP4F2 | IPI | UniProtKB:Q96KC8 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CYP4F2 | IPI | UniProtKB:Q96KR6 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CYP4F2 | IPI | UniProtKB:Q96MV1 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CYP4F2 | IPI | UniProtKB:Q96MV8 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CYP4F2 | IPI | UniProtKB:Q96QE2 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CYP4F2 | IPI | UniProtKB:Q9BRK0 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CYP4F2 | IPI | UniProtKB:Q9BZV2 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CYP4F2 | IPI | UniProtKB:Q9GZR5 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CYP4F2 | IPI | UniProtKB:Q9H2K0 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CYP4F2 | IPI | UniProtKB:Q9H6H4 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CYP4F2 | IPI | UniProtKB:Q9NPE6 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CYP4F2 | IPI | UniProtKB:Q9NW97 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CYP4F2 | IPI | UniProtKB:Q9NX18 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CYP4F2 | IPI | UniProtKB:Q9NY72 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CYP4F2 | IPI | UniProtKB:Q9Y282 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CYP4F2 | IPI | UniProtKB:Q9Y320 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CYP4F11 | IPI | UniProtKB:Q14696 | Human | PMID:32296183 |
| Molecular Function | GO:0052872 | tocotrienol omega-hydroxylase activity | CYP4F2 | IDA | | Human | PMID:20861217 |
| Cellular Component | GO:0016324 | apical plasma membrane | CYP4F2 | IDA | | Human | PMID:10660572 |
| Cellular Component | GO:0005737 | cytoplasm | CYP4F2 | IDA | | Human | PMID:10660572 |
| Cellular Component | GO:0043231 | intracellular membrane-bounded organelle | CYP4F2 | IDA | | Human | PMID:10660572 |
| Cellular Component | GO:0043231 | intracellular membrane-bounded organelle | CYP4F2 | IDA | | Human | PMID:20861217 |
| Cellular Component | GO:0043231 | intracellular membrane-bounded organelle | CYP4F2 | IDA | | Human | PMID:24138531 |
| Cellular Component | GO:0043231 | intracellular membrane-bounded organelle | CYP4F2 | IDA | | Human | PMID:9618440 |
| Cellular Component | GO:0043231 | intracellular membrane-bounded organelle | CYP4F2 | IDA | | Human | PMID:9799565 |
| Biological Process | GO:0019369 | arachidonic acid metabolic process | CYP4F2 | IDA | | Human | PMID:10660572 |
| Biological Process | GO:0019373 | epoxygenase P450 pathway | CYP4F2 | IDA | | Human | PMID:20861217 |
| Biological Process | GO:0019373 | epoxygenase P450 pathway | CYP4F2 | IDA | | Human | PMID:9618440 |
| Biological Process | GO:0006631 | fatty acid metabolic process | CYP4F11 | IDA | | Human | PMID:18065749 |
| Biological Process | GO:0010430 | fatty acid omega-oxidation | CYP4F2 | IDA | | Human | PMID:18577768 |
| Biological Process | GO:0036101 | leukotriene B4 catabolic process | CYP4F2 | IDA | | Human | PMID:10492403 |
| Biological Process | GO:0036101 | leukotriene B4 catabolic process | CYP4F2 | IDA | | Human | PMID:8026587 |
| Biological Process | GO:0036101 | leukotriene B4 catabolic process | CYP4F2 | IDA | | Human | PMID:9799565 |
| Biological Process | GO:0036101 | leukotriene B4 catabolic process | Cyp4f4 | IDA | | Rat | PMID:14634044|RGD:2301710 |
| Biological Process | GO:0036101 | leukotriene B4 catabolic process | Cyp4f4 | IDA | | Rat | PMID:9344476|RGD:15090848 |
| Biological Process | GO:0001676 | long-chain fatty acid metabolic process | CYP4F2 | IDA | | Human | PMID:18433732 |
| Biological Process | GO:0042361 | menaquinone catabolic process | CYP4F2 | IDA | | Human | PMID:24138531 |
| Biological Process | GO:0042361 | menaquinone catabolic process | CYP4F11 | IDA | | Human | PMID:24138531 |
| Biological Process | GO:0032304 | negative regulation of icosanoid secretion | CYP4F2 | IMP | | Human | PMID:18391101 |
| Biological Process | GO:0097267 | omega-hydroxylase P450 pathway | CYP4F2 | IDA | | Human | PMID:9799565 |
| Biological Process | GO:0097267 | omega-hydroxylase P450 pathway | CYP4F11 | IDA | | Human | PMID:15364545 |
| Biological Process | GO:0097267 | omega-hydroxylase P450 pathway | Cyp4f4 | IDA | | Rat | PMID:14634044|RGD:2301710 |
| Biological Process | GO:0042376 | phylloquinone catabolic process | CYP4F2 | IDA | | Human | PMID:24138531 |
| Biological Process | GO:0042376 | phylloquinone catabolic process | CYP4F11 | IDA | | Human | PMID:24138531 |
| Biological Process | GO:0032305 | positive regulation of icosanoid secretion | CYP4F2 | IMP | | Human | PMID:17341693 |
| Biological Process | GO:0003095 | pressure natriuresis | CYP4F2 | IEP | | Human | PMID:10660572 |
| Biological Process | GO:1905344 | prostaglandin catabolic process | Cyp4f4 | IDA | | Rat | PMID:9344476|RGD:15090848 |
| Biological Process | GO:0008217 | regulation of blood pressure | CYP4F2 | IMP | | Human | PMID:18391101 |
| Biological Process | GO:0003091 | renal water homeostasis | CYP4F2 | IEP | | Human | PMID:10660572 |
| Biological Process | GO:0055078 | sodium ion homeostasis | CYP4F2 | IEP | | Human | PMID:10660572 |
| Biological Process | GO:0000038 | very long-chain fatty acid metabolic process | CYP4F2 | IDA | | Human | PMID:18433732 |
| Biological Process | GO:0042360 | vitamin E metabolic process | CYP4F2 | IDA | | Human | PMID:20861217 |
| Biological Process | GO:0042377 | vitamin K catabolic process | CYP4F2 | IDA | | Human | PMID:19297519 |
| Biological Process | GO:0042377 | vitamin K catabolic process | CYP4F2 | IDA | | Human | PMID:24138531 |
| Biological Process | GO:0042377 | vitamin K catabolic process | CYP4F11 | IDA | | Human | PMID:24138531 |
| Biological Process | GO:0006805 | xenobiotic metabolic process | CYP4F2 | IMP | | Human | PMID:21084764 |
| Biological Process | GO:0006805 | xenobiotic metabolic process | CYP4F2 | IMP | | Human | PMID:21127708 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
 Analysis Tools
Analysis Tools