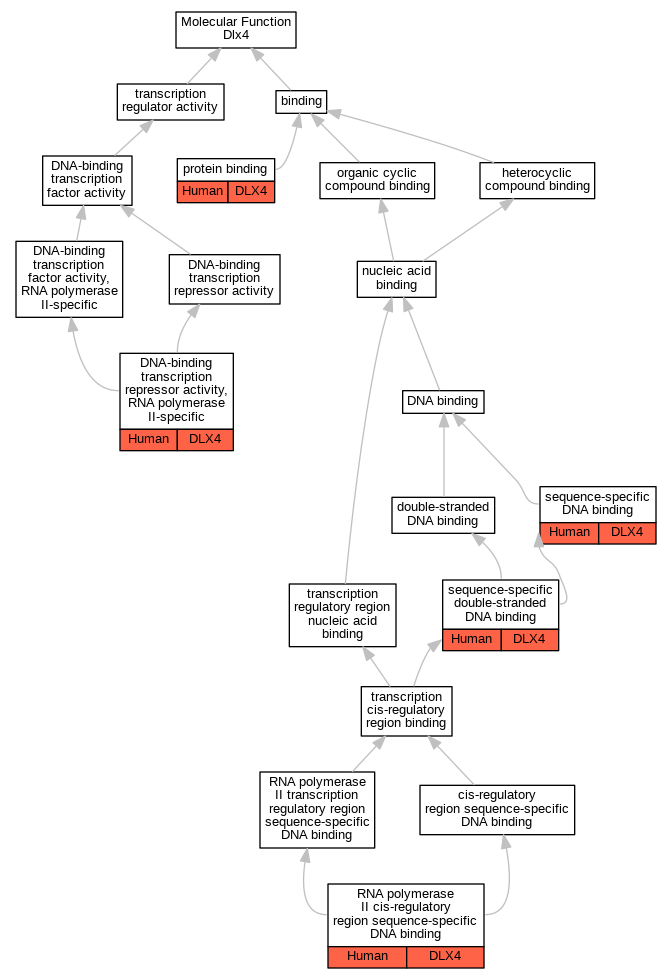
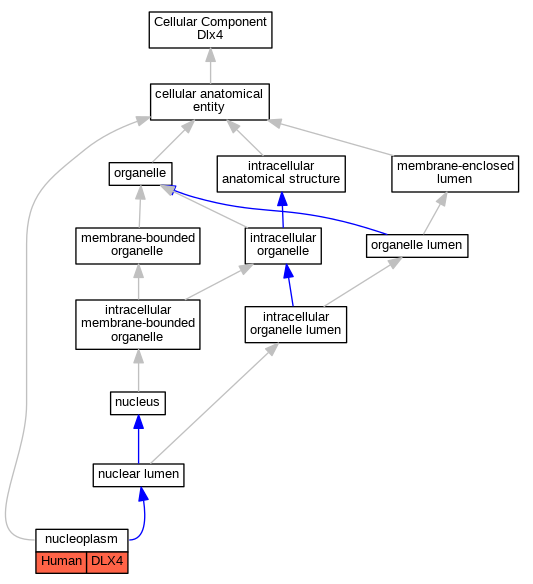
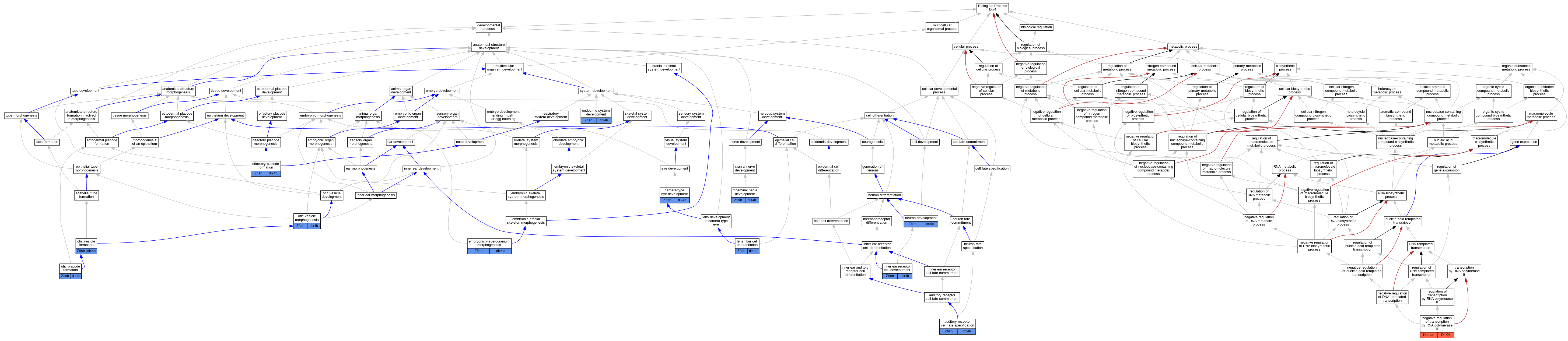
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0001227 | DNA-binding transcription repressor activity, RNA polymerase II-specific | DLX4 | IDA | | Human | PMID:11909945 |
| Molecular Function | GO:0005515 | protein binding | DLX4 | IPI | UniProtKB:P16333 | Human | PMID:17474147 |
| Molecular Function | GO:0005515 | protein binding | DLX4 | IPI | UniProtKB:P08047 | Human | PMID:21297662 |
| Molecular Function | GO:0005515 | protein binding | DLX4 | IPI | UniProtKB:Q13485 | Human | PMID:21297662 |
| Molecular Function | GO:0000978 | RNA polymerase II cis-regulatory region sequence-specific DNA binding | DLX4 | IDA | | Human | PMID:11909945 |
| Molecular Function | GO:0043565 | sequence-specific DNA binding | DLX4 | IDA | | Human | PMID:9073066 |
| Molecular Function | GO:1990837 | sequence-specific double-stranded DNA binding | DLX4 | IDA | | Human | PMID:28473536 |
| Cellular Component | GO:0005654 | nucleoplasm | DLX4 | IDA | | Human | GO_REF:0000052 |
| Biological Process | GO:0042667 | auditory receptor cell fate specification | dlx4b | IGI | ZFIN:ZDB-MRPHLNO-041109-1,ZFIN:ZDB-MRPHLNO-041110-1 | Zfish | PMID:23571216 |
| Biological Process | GO:0043010 | camera-type eye development | dlx4b | IGI | ZFIN:ZDB-MRPHLNO-041109-1,ZFIN:ZDB-MRPHLNO-041110-1 | Zfish | PMID:15728669 |
| Biological Process | GO:0048703 | embryonic viscerocranium morphogenesis | dlx4b | IGI | ZFIN:ZDB-MRPHLNO-041109-1,ZFIN:ZDB-MRPHLNO-050209-12,ZFIN:ZDB-MRPHLNO-060822-7 | Zfish | PMID:20573696 |
| Biological Process | GO:0048703 | embryonic viscerocranium morphogenesis | dlx4b | IGI | ZFIN:ZDB-GENO-100809-9,ZFIN:ZDB-MRPHLNO-041109-1,ZFIN:ZDB-MRPHLNO-050209-12,ZFIN:ZDB-MRPHLNO-060822-7 | Zfish | PMID:20573696 |
| Biological Process | GO:0048703 | embryonic viscerocranium morphogenesis | dlx4b | IMP | ZFIN:ZDB-MRPHLNO-050209-10,ZFIN:ZDB-MRPHLNO-050209-12 | Zfish | PMID:25954033 |
| Biological Process | GO:0035270 | endocrine system development | dlx4b | IGI | ZFIN:ZDB-MRPHLNO-041109-1,ZFIN:ZDB-MRPHLNO-041110-1 | Zfish | PMID:15728669 |
| Biological Process | GO:0060119 | inner ear receptor cell development | dlx4b | IGI | ZFIN:ZDB-GENO-180223-3 | Zfish | PMID:28751305 |
| Biological Process | GO:0070306 | lens fiber cell differentiation | dlx4b | IGI | ZFIN:ZDB-MRPHLNO-041109-1,ZFIN:ZDB-MRPHLNO-041110-1 | Zfish | PMID:15728669 |
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | DLX4 | IDA | | Human | PMID:11909945 |
| Biological Process | GO:0048666 | neuron development | dlx4b | IGI | ZFIN:ZDB-MRPHLNO-050209-11,ZFIN:ZDB-MRPHLNO-050209-12 | Zfish | PMID:15581883 |
| Biological Process | GO:0048666 | neuron development | dlx4b | IGI | ZFIN:ZDB-MRPHLNO-050209-10,ZFIN:ZDB-MRPHLNO-050209-9 | Zfish | PMID:15581883 |
| Biological Process | GO:0030910 | olfactory placode formation | dlx4b | IGI | ZFIN:ZDB-MRPHLNO-050208-13,ZFIN:ZDB-MRPHLNO-070504-2 | Zfish | PMID:12070088 |
| Biological Process | GO:0030910 | olfactory placode formation | dlx4b | IGI | ZFIN:ZDB-MRPHLNO-070504-1,ZFIN:ZDB-MRPHLNO-070504-3 | Zfish | PMID:12070088 |
| Biological Process | GO:0030910 | olfactory placode formation | dlx4b | IGI | ZFIN:ZDB-MRPHLNO-070504-1,ZFIN:ZDB-MRPHLNO-070504-2 | Zfish | PMID:12070088 |
| Biological Process | GO:0030910 | olfactory placode formation | dlx4b | IGI | ZFIN:ZDB-MRPHLNO-050208-13,ZFIN:ZDB-MRPHLNO-070504-3 | Zfish | PMID:12070088 |
| Biological Process | GO:0043049 | otic placode formation | dlx4b | IGI | ZFIN:ZDB-MRPHLNO-050208-13,ZFIN:ZDB-MRPHLNO-070504-3 | Zfish | PMID:12070088 |
| Biological Process | GO:0043049 | otic placode formation | dlx4b | IGI | ZFIN:ZDB-MRPHLNO-070504-1,ZFIN:ZDB-MRPHLNO-070504-2 | Zfish | PMID:12070088 |
| Biological Process | GO:0043049 | otic placode formation | dlx4b | IGI | ZFIN:ZDB-GENE-980526-280 | Zfish | PMID:15188428 |
| Biological Process | GO:0043049 | otic placode formation | dlx4b | IGI | ZFIN:ZDB-GENE-001103-1,ZFIN:ZDB-GENE-980526-280 | Zfish | PMID:12668634 |
| Biological Process | GO:0043049 | otic placode formation | dlx4b | IGI | ZFIN:ZDB-MRPHLNO-070504-1,ZFIN:ZDB-MRPHLNO-070504-3 | Zfish | PMID:12070088 |
| Biological Process | GO:0043049 | otic placode formation | dlx4b | IGI | ZFIN:ZDB-MRPHLNO-050208-13,ZFIN:ZDB-MRPHLNO-070504-2 | Zfish | PMID:12070088 |
| Biological Process | GO:0030916 | otic vesicle formation | dlx4b | IGI | ZFIN:ZDB-GENE-980526-280,ZFIN:ZDB-GENO-030617-1 | Zfish | PMID:15188428 |
| Biological Process | GO:0071600 | otic vesicle morphogenesis | dlx4b | IGI | ZFIN:ZDB-GENO-180223-3 | Zfish | PMID:28751305 |
| Biological Process | GO:0021559 | trigeminal nerve development | dlx4b | IGI | ZFIN:ZDB-MRPHLNO-050209-11,ZFIN:ZDB-MRPHLNO-050209-12 | Zfish | PMID:15581883 |
| Biological Process | GO:0021559 | trigeminal nerve development | dlx4b | IGI | ZFIN:ZDB-MRPHLNO-050209-10,ZFIN:ZDB-MRPHLNO-050209-9 | Zfish | PMID:15581883 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
 Analysis Tools
Analysis Tools