|
Source |
Alliance of Genome Resources |
| Category | ID | Classification term | Gene | Evidence | Inferred from | Organism | Reference |
|---|---|---|---|---|---|---|---|
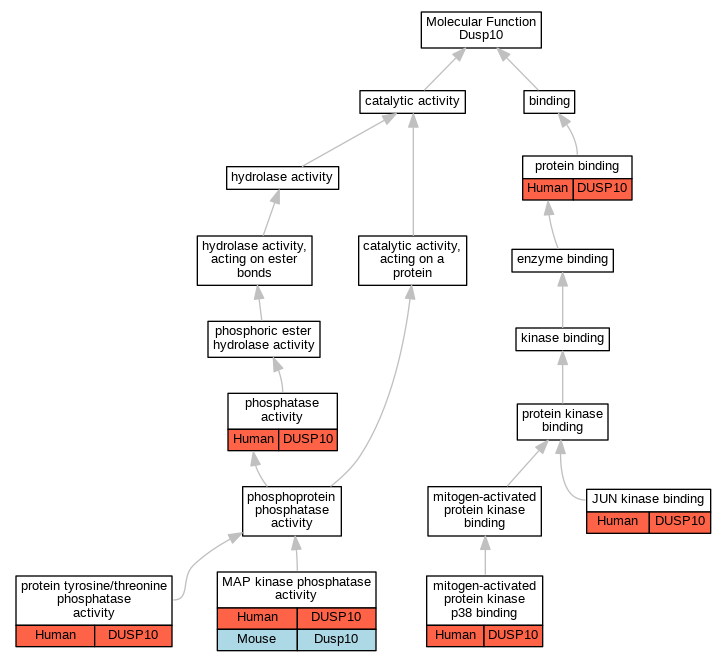
| Molecular Function | GO:0008432 | JUN kinase binding | DUSP10 | IDA | Human | PMID:10391943 | |
| Molecular Function | GO:0033549 | MAP kinase phosphatase activity | DUSP10 | IDA | Human | PMID:22375048 | |
| Molecular Function | GO:0033549 | MAP kinase phosphatase activity | Dusp10 | IDA | Mouse | MGI:MGI:5905244|PMID:25772234 | |
| Molecular Function | GO:0048273 | mitogen-activated protein kinase p38 binding | DUSP10 | IPI | UniProtKB:Q16539 | Human | PMID:10391943 |
| Molecular Function | GO:0016791 | phosphatase activity | DUSP10 | IDA | Human | PMID:16806267 | |
| Molecular Function | GO:0016791 | phosphatase activity | DUSP10 | IDA | Human | PMID:24531476 | |
| Molecular Function | GO:0005515 | protein binding | DUSP10 | IPI | UniProtKB:P45984 | Human | PMID:16288922 |
| Molecular Function | GO:0005515 | protein binding | DUSP10 | IPI | UniProtKB:P45984 | Human | PMID:27880917 |
| Molecular Function | GO:0005515 | protein binding | DUSP10 | IPI | UniProtKB:P45983-4 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | DUSP10 | IPI | UniProtKB:P45984 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | DUSP10 | IPI | UniProtKB:P49639 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | DUSP10 | IPI | UniProtKB:Q53EP0-3 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | DUSP10 | IPI | UniProtKB:Q9BYR7 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | DUSP10 | IPI | UniProtKB:P01112 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | DUSP10 | IPI | UniProtKB:P02489 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | DUSP10 | IPI | UniProtKB:P06396 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | DUSP10 | IPI | UniProtKB:P22607 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | DUSP10 | IPI | UniProtKB:Q14204 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | DUSP10 | IPI | UniProtKB:Q14957 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | DUSP10 | IPI | UniProtKB:Q86V38 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | DUSP10 | IPI | UniProtKB:Q9UMX0 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | DUSP10 | IPI | UniProtKB:Q9Y5Z0 | Human | PMID:32814053 |
| Molecular Function | GO:0008330 | protein tyrosine/threonine phosphatase activity | DUSP10 | IDA | Human | PMID:10391943 | |
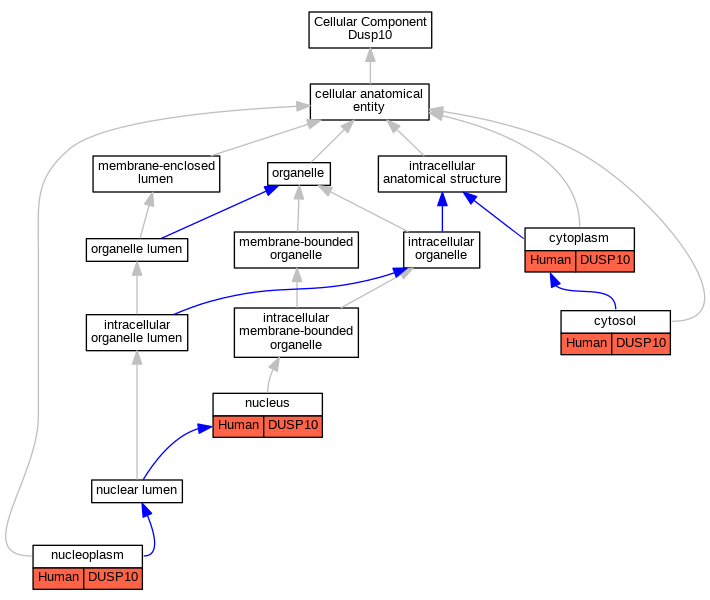
| Cellular Component | GO:0005737 | cytoplasm | DUSP10 | IDA | Human | PMID:10391943 | |
| Cellular Component | GO:0005829 | cytosol | DUSP10 | IDA | Human | GO_REF:0000052 | |
| Cellular Component | GO:0005654 | nucleoplasm | DUSP10 | IDA | Human | GO_REF:0000052 | |
| Cellular Component | GO:0005634 | nucleus | DUSP10 | IDA | Human | PMID:10391943 | |
| Biological Process | GO:0016311 | dephosphorylation | DUSP10 | IDA | Human | PMID:24531476 | |
| Biological Process | GO:0010633 | negative regulation of epithelial cell migration | Dusp10 | IMP | Mouse | MGI:MGI:5905244|PMID:25772234 | |
| Biological Process | GO:0050680 | negative regulation of epithelial cell proliferation | Dusp10 | IMP | Mouse | MGI:MGI:5905244|PMID:25772234 | |
| Biological Process | GO:1905042 | negative regulation of epithelium regeneration | Dusp10 | IDA | Mouse | MGI:MGI:5905244|PMID:25772234 | |
| Biological Process | GO:0070373 | negative regulation of ERK1 and ERK2 cascade | Dusp10 | IMP | Mouse | MGI:MGI:5905244|PMID:25772234 | |
| Biological Process | GO:0046329 | negative regulation of JNK cascade | DUSP10 | IDA | Human | PMID:10391943 | |
| Biological Process | GO:0046329 | negative regulation of JNK cascade | Dusp10 | IMP | MGI:MGI:3052343 | Mouse | PMID:15306813 |
| Biological Process | GO:0043508 | negative regulation of JUN kinase activity | DUSP10 | IDA | Human | PMID:10391943 | |
| Biological Process | GO:0043508 | negative regulation of JUN kinase activity | Dusp10 | IMP | MGI:MGI:3052343 | Mouse | PMID:15306813 |
| Biological Process | GO:0048715 | negative regulation of oligodendrocyte differentiation | Dusp10 | IMP | Rat | PMID:19139271|RGD:7775012 | |
| Biological Process | GO:1903753 | negative regulation of p38MAPK cascade | DUSP10 | IDA | Human | PMID:10391943 | |
| Biological Process | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation | Dusp10 | IDA | Mouse | PMID:23977283 | |
| Biological Process | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response | Dusp10 | IMP | MGI:MGI:3052343 | Mouse | PMID:19696743 |
| Biological Process | GO:0032873 | negative regulation of stress-activated MAPK cascade | Dusp10 | IMP | MGI:MGI:3052343 | Mouse | PMID:19696743 |
| Biological Process | GO:0048709 | oligodendrocyte differentiation | Dusp10 | IMP | Mouse | PMID:19139271 | |
| Biological Process | GO:0035970 | peptidyl-threonine dephosphorylation | DUSP10 | IDA | Human | PMID:10391943 | |
| Biological Process | GO:1990264 | peptidyl-tyrosine dephosphorylation involved in inactivation of protein kinase activity | DUSP10 | IDA | Human | PMID:10391943 | |
| Biological Process | GO:0045591 | positive regulation of regulatory T cell differentiation | DUSP10 | IMP | Human | PMID:22387553 | |
| Biological Process | GO:0006470 | protein dephosphorylation | Dusp10 | IDA | Mouse | MGI:MGI:5905244|PMID:25772234 | |
| Biological Process | GO:0002819 | regulation of adaptive immune response | Dusp10 | IMP | MGI:MGI:3052343 | Mouse | PMID:15306813 |
| Biological Process | GO:0090335 | regulation of brown fat cell differentiation | Dusp10 | IDA | Mouse | PMID:23874687 | |
| Biological Process | GO:0045088 | regulation of innate immune response | Dusp10 | IMP | MGI:MGI:3052343 | Mouse | PMID:15306813 |
| Biological Process | GO:0032496 | response to lipopolysaccharide | Dusp10 | IMP | MGI:MGI:3052343 | Mouse | PMID:19696743 |
| Biological Process | GO:0010033 | response to organic substance | Dusp10 | IEP | Rat | PMID:11027531|RGD:2301725 | |
| Biological Process | GO:0051403 | stress-activated MAPK cascade | Dusp10 | IMP | MGI:MGI:3052343 | Mouse | PMID:19696743 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 06/12/2024 MGI 6.13 |

|
|
|
||