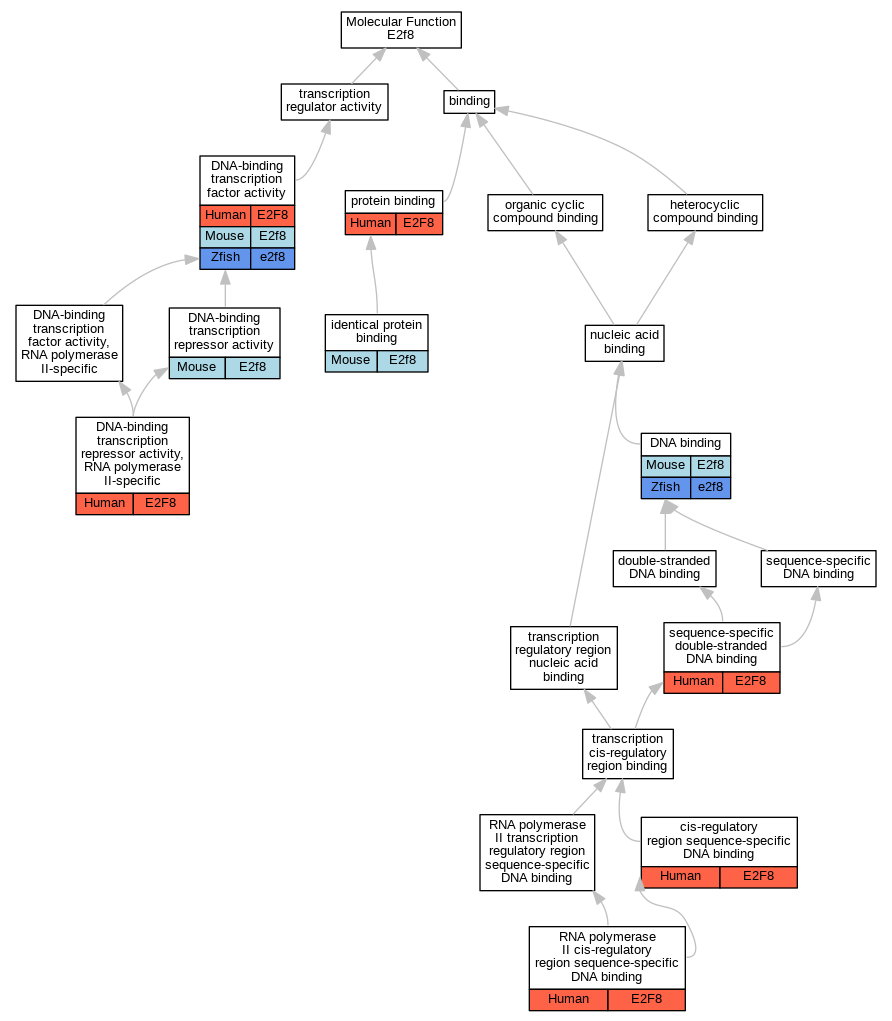
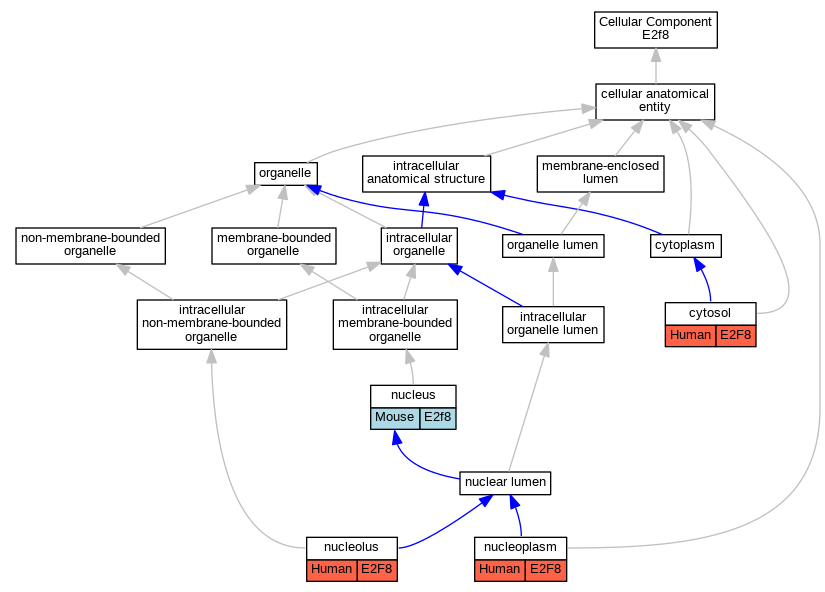
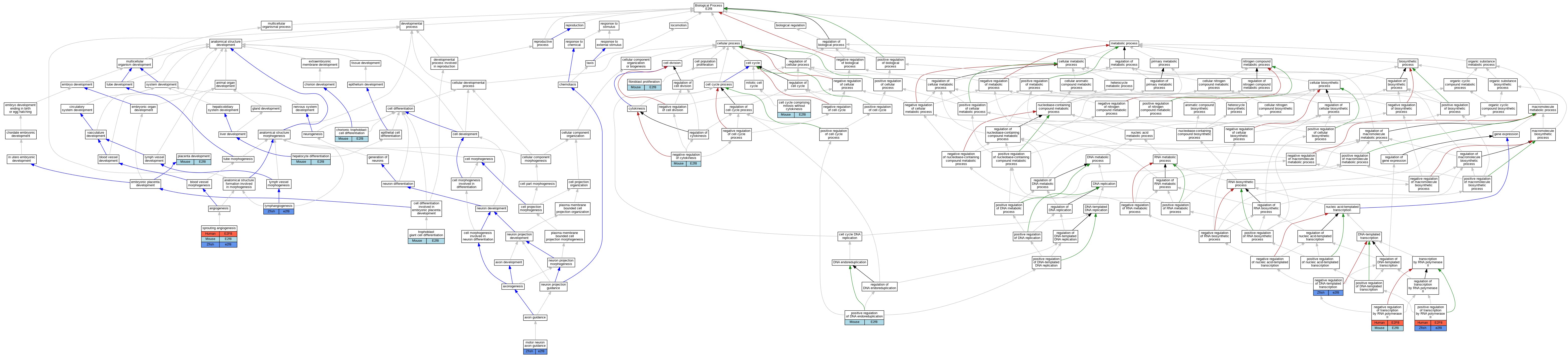
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0000987 | cis-regulatory region sequence-specific DNA binding | E2F8 | IDA | | Human | PMID:22903062 |
| Molecular Function | GO:0003677 | DNA binding | E2f8 | IDA | | Mouse | PMID:15722552 |
| Molecular Function | GO:0003677 | DNA binding | e2f8 | IDA | | Zfish | PMID:22903062 |
| Molecular Function | GO:0003700 | DNA-binding transcription factor activity | E2F8 | IMP | | Human | PMID:22903062 |
| Molecular Function | GO:0003700 | DNA-binding transcription factor activity | E2f8 | IMP | | Mouse | MGI:MGI:5449497|PMID:23064266 |
| Molecular Function | GO:0003700 | DNA-binding transcription factor activity | E2f8 | IMP | | Mouse | MGI:MGI:5449498|PMID:23064264 |
| Molecular Function | GO:0003700 | DNA-binding transcription factor activity | E2f8 | IMP | | Mouse | MGI:MGI:5319718|PMID:22516201 |
| Molecular Function | GO:0003700 | DNA-binding transcription factor activity | e2f8 | IMP | | Zfish | PMID:22903062|ZFIN:ZDB-PUB-120822-27 |
| Molecular Function | GO:0001217 | DNA-binding transcription repressor activity | E2f8 | IMP | | Mouse | MGI:MGI:5319718|PMID:22516201 |
| Molecular Function | GO:0001217 | DNA-binding transcription repressor activity | E2f8 | IMP | | Mouse | MGI:MGI:5449498|PMID:23064264 |
| Molecular Function | GO:0001217 | DNA-binding transcription repressor activity | E2f8 | IMP | | Mouse | MGI:MGI:5449497|PMID:23064266 |
| Molecular Function | GO:0001227 | DNA-binding transcription repressor activity, RNA polymerase II-specific | E2F8 | IDA | | Human | PMID:16179649 |
| Molecular Function | GO:0042802 | identical protein binding | E2f8 | IPI | PR:Q58FA4 | Mouse | PMID:15722552 |
| Molecular Function | GO:0005515 | protein binding | E2F8 | IPI | UniProtKB:Q96AV8 | Human | PMID:18202719 |
| Molecular Function | GO:0005515 | protein binding | E2F8 | IPI | UniProtKB:Q12800 | Human | PMID:25416956 |
| Molecular Function | GO:0005515 | protein binding | E2F8 | IPI | UniProtKB:Q13643 | Human | PMID:25416956 |
| Molecular Function | GO:0005515 | protein binding | E2F8 | IPI | UniProtKB:Q6FHY5 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | E2F8 | IPI | UniProtKB:Q9UNE7 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | E2F8 | IPI | UniProtKB:O14645 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | E2F8 | IPI | UniProtKB:O14901 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | E2F8 | IPI | UniProtKB:P01112 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | E2F8 | IPI | UniProtKB:P06396 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | E2F8 | IPI | UniProtKB:P07550 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | E2F8 | IPI | UniProtKB:P22607 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | E2F8 | IPI | UniProtKB:P37840 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | E2F8 | IPI | UniProtKB:P42858 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | E2F8 | IPI | UniProtKB:P46379-2 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | E2F8 | IPI | UniProtKB:P55212 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | E2F8 | IPI | UniProtKB:Q14957 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | E2F8 | IPI | UniProtKB:Q7Z412 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | E2F8 | IPI | UniProtKB:Q92870-2 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | E2F8 | IPI | UniProtKB:Q9UMX0 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | E2F8 | IPI | UniProtKB:Q9Y649 | Human | PMID:32814053 |
| Molecular Function | GO:0000978 | RNA polymerase II cis-regulatory region sequence-specific DNA binding | E2F8 | IDA | | Human | PMID:16179649 |
| Molecular Function | GO:1990837 | sequence-specific double-stranded DNA binding | E2F8 | IDA | | Human | PMID:28473536 |
| Cellular Component | GO:0005829 | cytosol | E2F8 | IDA | | Human | GO_REF:0000052 |
| Cellular Component | GO:0005730 | nucleolus | E2F8 | IDA | | Human | GO_REF:0000052 |
| Cellular Component | GO:0005654 | nucleoplasm | E2F8 | IDA | | Human | GO_REF:0000052 |
| Cellular Component | GO:0005634 | nucleus | E2f8 | IDA | | Mouse | PMID:15722552 |
| Biological Process | GO:0033301 | cell cycle comprising mitosis without cytokinesis | E2f8 | IMP | | Mouse | MGI:MGI:5449497|PMID:23064266 |
| Biological Process | GO:0060718 | chorionic trophoblast cell differentiation | E2f8 | IMP | | Mouse | MGI:MGI:5319718|PMID:22516201 |
| Biological Process | GO:0048144 | fibroblast proliferation | E2f8 | IDA | | Mouse | PMID:15722552 |
| Biological Process | GO:0070365 | hepatocyte differentiation | E2f8 | IMP | | Mouse | MGI:MGI:5449497|PMID:23064266 |
| Biological Process | GO:0070365 | hepatocyte differentiation | E2f8 | IMP | | Mouse | MGI:MGI:5449498|PMID:23064264 |
| Biological Process | GO:0001946 | lymphangiogenesis | e2f8 | IGI | ZFIN:ZDB-MRPHLNO-121116-1,ZFIN:ZDB-MRPHLNO-121116-2 | Zfish | PMID:24069224 |
| Biological Process | GO:0001946 | lymphangiogenesis | e2f8 | IGI | ZFIN:ZDB-MRPHLNO-090507-1,ZFIN:ZDB-MRPHLNO-121116-1,ZFIN:ZDB-MRPHLNO-121116-2 | Zfish | PMID:24069224 |
| Biological Process | GO:0001946 | lymphangiogenesis | e2f8 | IGI | ZFIN:ZDB-MRPHLNO-100205-5,ZFIN:ZDB-MRPHLNO-121116-1,ZFIN:ZDB-MRPHLNO-121116-2 | Zfish | PMID:24069224 |
| Biological Process | GO:0008045 | motor neuron axon guidance | e2f8 | IGI | ZFIN:ZDB-GENE-030131-3527 | Zfish | PMID:26681691 |
| Biological Process | GO:0008045 | motor neuron axon guidance | e2f8 | IGI | ZFIN:ZDB-GENE-030131-3527,ZFIN:ZDB-GENE-030519-2 | Zfish | PMID:26681691 |
| Biological Process | GO:0032466 | negative regulation of cytokinesis | E2f8 | IMP | | Mouse | MGI:MGI:5449497|PMID:23064266 |
| Biological Process | GO:0032466 | negative regulation of cytokinesis | E2f8 | IMP | | Mouse | MGI:MGI:5449498|PMID:23064264 |
| Biological Process | GO:0045892 | negative regulation of DNA-templated transcription | e2f8 | IGI | ZFIN:ZDB-GENE-030131-3527 | Zfish | PMID:26681691 |
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | E2F8 | IDA | | Human | PMID:16179649 |
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | E2f8 | IMP | | Mouse | MGI:MGI:5449497|PMID:23064266 |
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | E2f8 | IMP | | Mouse | MGI:MGI:5319718|PMID:22516201 |
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | E2f8 | IMP | | Mouse | MGI:MGI:5449498|PMID:23064264 |
| Biological Process | GO:0001890 | placenta development | E2f8 | IMP | | Mouse | MGI:MGI:5319718|PMID:22516201 |
| Biological Process | GO:0032877 | positive regulation of DNA endoreduplication | E2f8 | IMP | | Mouse | MGI:MGI:5449497|PMID:23064266 |
| Biological Process | GO:0032877 | positive regulation of DNA endoreduplication | E2f8 | IMP | | Mouse | MGI:MGI:5449498|PMID:23064264 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | E2F8 | IMP | | Human | PMID:22903062 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | e2f8 | IMP | | Zfish | PMID:22903062|ZFIN:ZDB-PUB-120822-27 |
| Biological Process | GO:0002040 | sprouting angiogenesis | E2F8 | IMP | | Human | PMID:22903062 |
| Biological Process | GO:0002040 | sprouting angiogenesis | E2f8 | IMP | | Mouse | MGI:MGI:5447251|PMID:22903062 |
| Biological Process | GO:0002040 | sprouting angiogenesis | e2f8 | IMP | | Zfish | PMID:22903062|ZFIN:ZDB-PUB-120822-27 |
| Biological Process | GO:0002040 | sprouting angiogenesis | e2f8 | IGI | ZFIN:ZDB-MRPHLNO-121116-1,ZFIN:ZDB-MRPHLNO-121116-2 | Zfish | PMID:24069224 |
| Biological Process | GO:0002040 | sprouting angiogenesis | e2f8 | IGI | ZFIN:ZDB-MRPHLNO-090507-1,ZFIN:ZDB-MRPHLNO-121116-1,ZFIN:ZDB-MRPHLNO-121116-2 | Zfish | PMID:24069224 |
| Biological Process | GO:0002040 | sprouting angiogenesis | e2f8 | IGI | ZFIN:ZDB-MRPHLNO-100205-5,ZFIN:ZDB-MRPHLNO-121116-1,ZFIN:ZDB-MRPHLNO-121116-2 | Zfish | PMID:24069224 |
| Biological Process | GO:0002040 | sprouting angiogenesis | e2f8 | IGI | ZFIN:ZDB-MRPHLNO-121116-1,ZFIN:ZDB-MRPHLNO-121116-2 | Zfish | PMID:22903062 |
| Biological Process | GO:0002040 | sprouting angiogenesis | e2f8 | IGI | ZFIN:ZDB-MRPHLNO-050513-12,ZFIN:ZDB-MRPHLNO-121116-1,ZFIN:ZDB-MRPHLNO-121116-2 | Zfish | PMID:22903062 |
| Biological Process | GO:0060707 | trophoblast giant cell differentiation | E2f8 | IMP | | Mouse | MGI:MGI:5449497|PMID:23064266 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
 Analysis Tools
Analysis Tools