| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
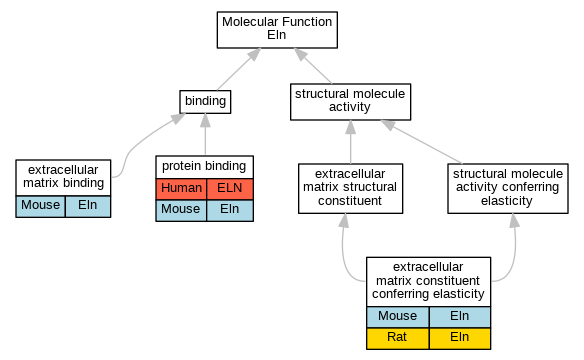
| Molecular Function | GO:0050840 | extracellular matrix binding | Eln | IDA | | Mouse | PMID:18757743 |
| Molecular Function | GO:0030023 | extracellular matrix constituent conferring elasticity | Eln | IDA | | Mouse | MGI:MGI:5142190|PMID:20576933 |
| Molecular Function | GO:0030023 | extracellular matrix constituent conferring elasticity | Eln | IMP | | Rat | PMID:10090127|RGD:731206 |
| Molecular Function | GO:0005515 | protein binding | ELN | IPI | UniProtKB:Q9WVH8 | Human | PMID:11805834 |
| Molecular Function | GO:0005515 | protein binding | ELN | IPI | UniProtKB:P35555 | Human | PMID:15790312 |
| Molecular Function | GO:0005515 | protein binding | ELN | IPI | UniProtKB:Q9UBX5 | Human | PMID:15790312 |
| Molecular Function | GO:0005515 | protein binding | ELN | IPI | UniProtKB:O95967 | Human | PMID:16478991 |
| Molecular Function | GO:0005515 | protein binding | ELN | IPI | UniProtKB:Q9UBX5 | Human | PMID:17035250 |
| Molecular Function | GO:0005515 | protein binding | ELN | IPI | UniProtKB:Q9WVJ9 | Human | PMID:17324935 |
| Molecular Function | GO:0005515 | protein binding | ELN | IPI | UniProtKB:O95967 | Human | PMID:19570982 |
| Molecular Function | GO:0005515 | protein binding | ELN | IPI | UniProtKB:P28300 | Human | PMID:19570982 |
| Molecular Function | GO:0005515 | protein binding | ELN | IPI | UniProtKB:Q9UBX5 | Human | PMID:19570982 |
| Molecular Function | GO:0005515 | protein binding | Eln | IPI | PR:P97873 | Mouse | PMID:16251195 |
| Molecular Function | GO:0005515 | protein binding | Eln | IPI | PR:P28301 | Mouse | PMID:16251195 |
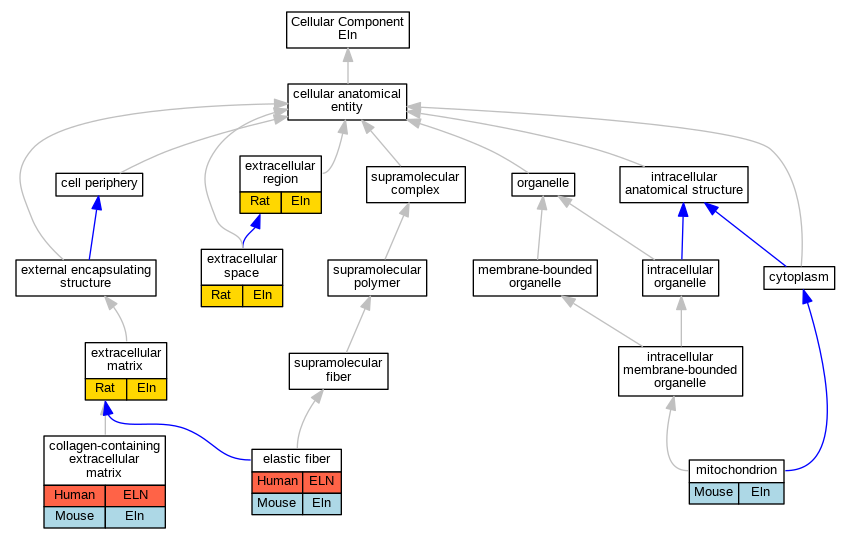
| Cellular Component | GO:0062023 | collagen-containing extracellular matrix | ELN | HDA | | Human | PMID:27559042 |
| Cellular Component | GO:0062023 | collagen-containing extracellular matrix | ELN | HDA | | Human | PMID:28327460 |
| Cellular Component | GO:0062023 | collagen-containing extracellular matrix | ELN | HDA | | Human | PMID:25037231 |
| Cellular Component | GO:0062023 | collagen-containing extracellular matrix | ELN | HDA | | Human | PMID:28675934 |
| Cellular Component | GO:0062023 | collagen-containing extracellular matrix | Eln | HDA | | Mouse | MGI:MGI:6201834|PMID:22159717 |
| Cellular Component | GO:0062023 | collagen-containing extracellular matrix | Eln | HDA | | Mouse | MGI:MGI:6201853|PMID:28071719 |
| Cellular Component | GO:0071953 | elastic fiber | ELN | IDA | | Human | PMID:10424889 |
| Cellular Component | GO:0071953 | elastic fiber | ELN | IDA | | Human | PMID:7534784 |
| Cellular Component | GO:0071953 | elastic fiber | Eln | IDA | | Mouse | PMID:17099216 |
| Cellular Component | GO:0031012 | extracellular matrix | Eln | IDA | | Rat | PMID:18847363|RGD:9585685 |
| Cellular Component | GO:0005576 | extracellular region | Eln | IDA | | Rat | PMID:16473861|RGD:1601028 |
| Cellular Component | GO:0005615 | extracellular space | Eln | IDA | | Rat | PMID:8238530|RGD:9585669 |
| Cellular Component | GO:0005739 | mitochondrion | Eln | HDA | | Mouse | PMID:18614015 |
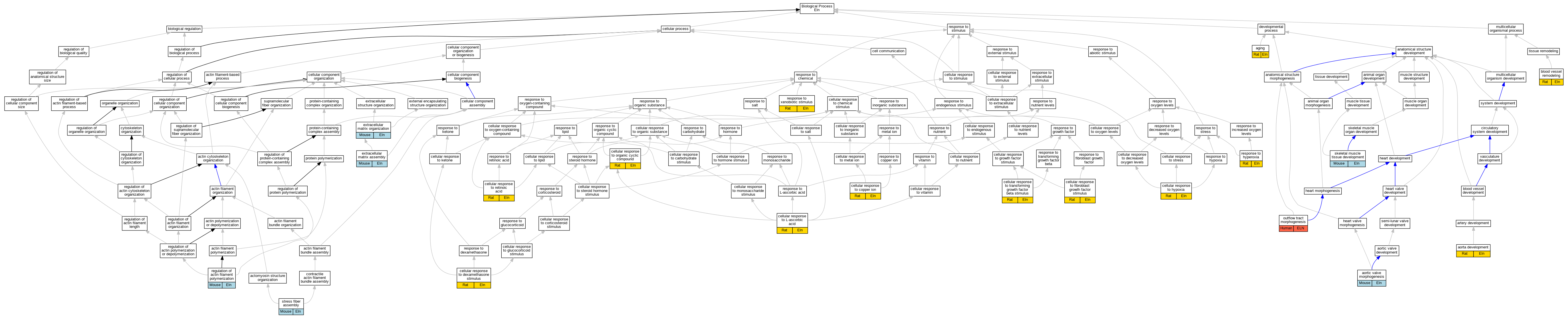
| Biological Process | GO:0007568 | aging | Eln | IEP | | Rat | PMID:23354684|RGD:9585655 |
| Biological Process | GO:0035904 | aorta development | Eln | IEP | | Rat | PMID:18803461|RGD:7394730 |
| Biological Process | GO:0003180 | aortic valve morphogenesis | Eln | IMP | | Mouse | MGI:MGI:5142190|PMID:20576933 |
| Biological Process | GO:0001974 | blood vessel remodeling | Eln | IDA | | Rat | PMID:12844513|RGD:1580324 |
| Biological Process | GO:0071280 | cellular response to copper ion | Eln | IEP | | Rat | PMID:18847363|RGD:9585685 |
| Biological Process | GO:0071549 | cellular response to dexamethasone stimulus | Eln | IEP | | Rat | PMID:2235137|RGD:9585723 |
| Biological Process | GO:0044344 | cellular response to fibroblast growth factor stimulus | Eln | IEP | | Rat | PMID:11967999|RGD:9585657 |
| Biological Process | GO:0071456 | cellular response to hypoxia | Eln | IEP | | Rat | PMID:16055482|RGD:9585727 |
| Biological Process | GO:0071298 | cellular response to L-ascorbic acid | Eln | IEP | | Rat | PMID:2859894|RGD:9585689 |
| Biological Process | GO:0071407 | cellular response to organic cyclic compound | Eln | IEP | | Rat | PMID:8997264|RGD:9585728 |
| Biological Process | GO:0071300 | cellular response to retinoic acid | Eln | IEP | | Rat | PMID:8238530|RGD:9585669 |
| Biological Process | GO:0071560 | cellular response to transforming growth factor beta stimulus | Eln | IEP | | Rat | PMID:9224206|RGD:9585659 |
| Biological Process | GO:0085029 | extracellular matrix assembly | Eln | IMP | | Mouse | MGI:MGI:5142190|PMID:20576933 |
| Biological Process | GO:0030198 | extracellular matrix organization | Eln | IDA | | Mouse | PMID:18757743 |
| Biological Process | GO:0003151 | outflow tract morphogenesis | ELN | IMP | | Human | PMID:8132745 |
| Biological Process | GO:0030833 | regulation of actin filament polymerization | Eln | IMP | MGI:MGI:2153007 | Mouse | PMID:14614988 |
| Biological Process | GO:0055093 | response to hyperoxia | Eln | IEP | | Rat | PMID:9609733|RGD:9585726 |
| Biological Process | GO:0009410 | response to xenobiotic stimulus | Eln | IEP | | Rat | PMID:18279932|RGD:9585725 |
| Biological Process | GO:0007519 | skeletal muscle tissue development | Eln | IMP | MGI:MGI:2153007 | Mouse | PMID:14614988 |
| Biological Process | GO:0043149 | stress fiber assembly | Eln | IMP | MGI:MGI:2153007 | Mouse | PMID:14614988 |
 Analysis Tools
Analysis Tools