|
Source |
Alliance of Genome Resources |
| Category | ID | Classification term | Gene | Evidence | Inferred from | Organism | Reference |
|---|---|---|---|---|---|---|---|
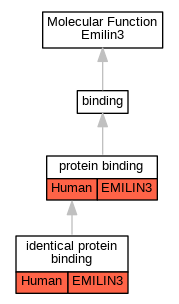
| Molecular Function | GO:0042802 | identical protein binding | EMILIN3 | IPI | UniProtKB:Q9NT22 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | EMILIN3 | IPI | UniProtKB:O00560 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | EMILIN3 | IPI | UniProtKB:O75934 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | EMILIN3 | IPI | UniProtKB:Q2TAC2-2 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | EMILIN3 | IPI | UniProtKB:Q96HA8 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | EMILIN3 | IPI | UniProtKB:Q9H8Y8 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | EMILIN3 | IPI | UniProtKB:Q9NVF7 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | EMILIN3 | IPI | UniProtKB:Q9Y316 | Human | PMID:32296183 |
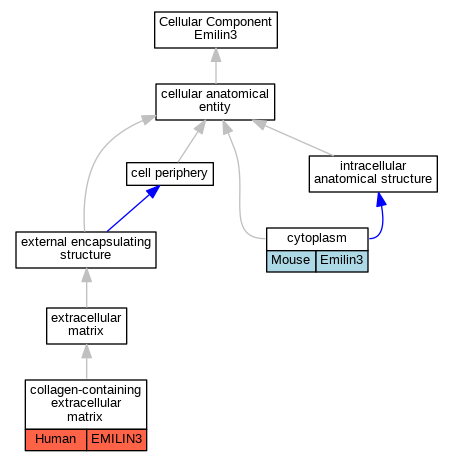
| Cellular Component | GO:0062023 | collagen-containing extracellular matrix | EMILIN3 | HDA | Human | PMID:25037231 | |
| Cellular Component | GO:0005737 | cytoplasm | Emilin3 | IDA | Mouse | PMID:14706625 | |
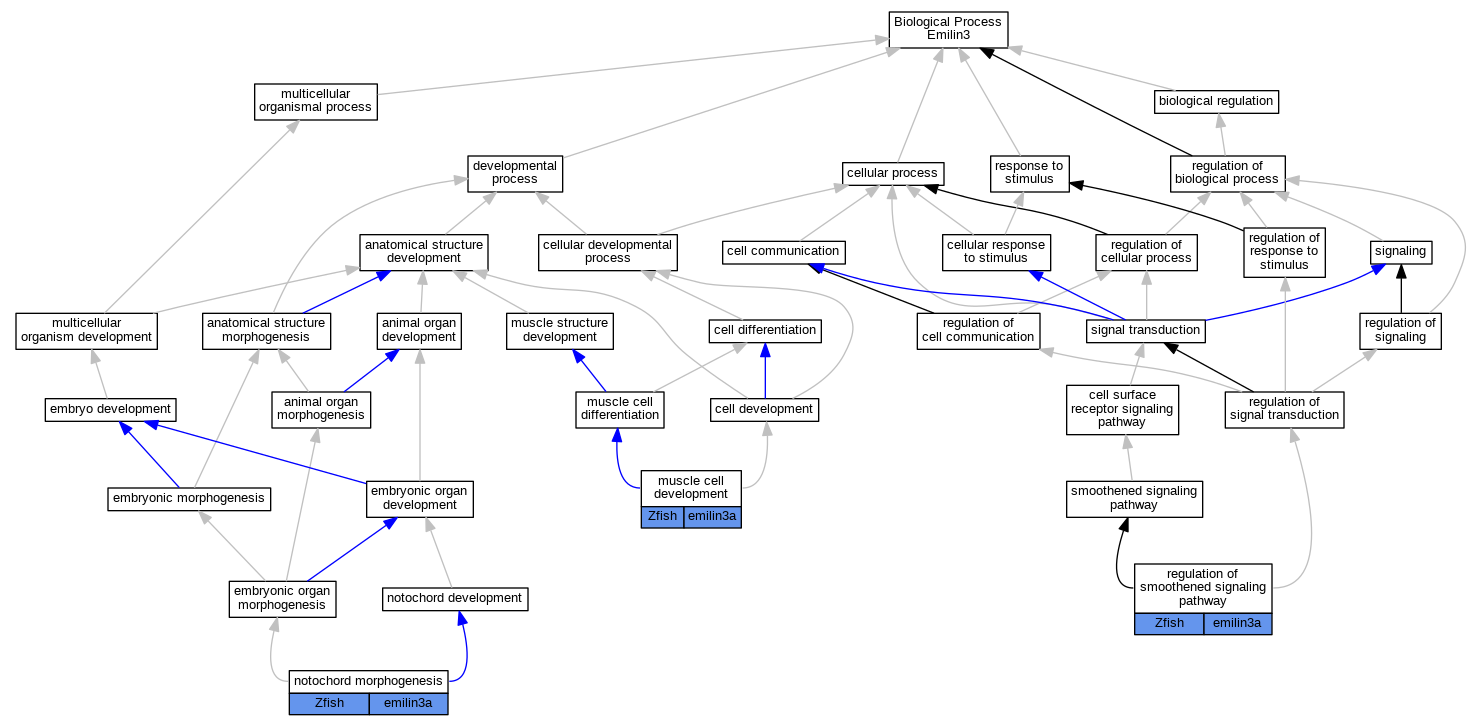
| Biological Process | GO:0055001 | muscle cell development | emilin3a | IGI | ZFIN:ZDB-MRPHLNO-140114-6,ZFIN:ZDB-MRPHLNO-140114-7 | Zfish | PMID:24131633 |
| Biological Process | GO:0055001 | muscle cell development | emilin3a | IGI | ZFIN:ZDB-MRPHLNO-050510-1,ZFIN:ZDB-MRPHLNO-140114-6,ZFIN:ZDB-MRPHLNO-140114-7 | Zfish | PMID:24131633 |
| Biological Process | GO:0048570 | notochord morphogenesis | emilin3a | IGI | ZFIN:ZDB-MRPHLNO-140114-3,ZFIN:ZDB-MRPHLNO-140114-5 | Zfish | PMID:24131633 |
| Biological Process | GO:0048570 | notochord morphogenesis | emilin3a | IGI | ZFIN:ZDB-MRPHLNO-140114-6,ZFIN:ZDB-MRPHLNO-140114-7 | Zfish | PMID:24131633 |
| Biological Process | GO:0008589 | regulation of smoothened signaling pathway | emilin3a | IGI | ZFIN:ZDB-MRPHLNO-140114-6,ZFIN:ZDB-MRPHLNO-140114-7 | Zfish | PMID:24131633 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 07/05/2024 MGI 6.24 |

|
|
|
||