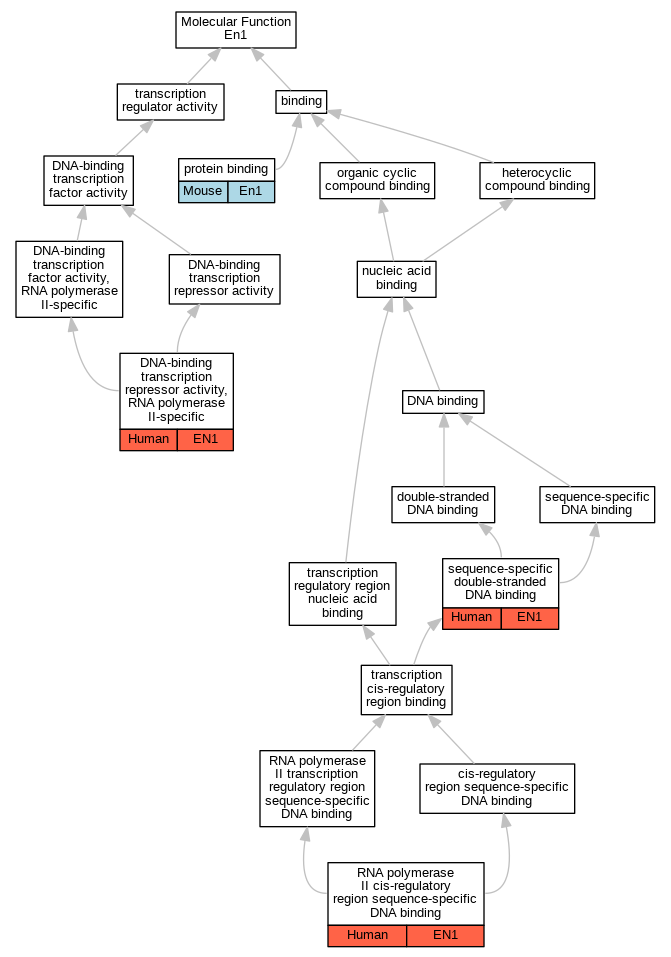
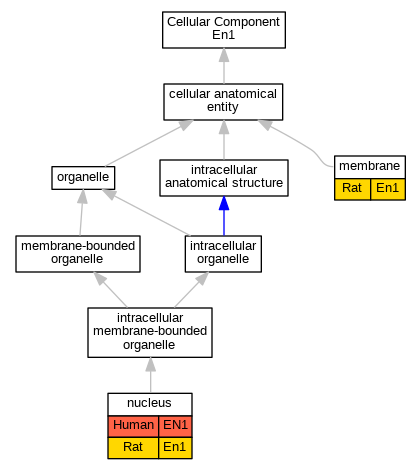
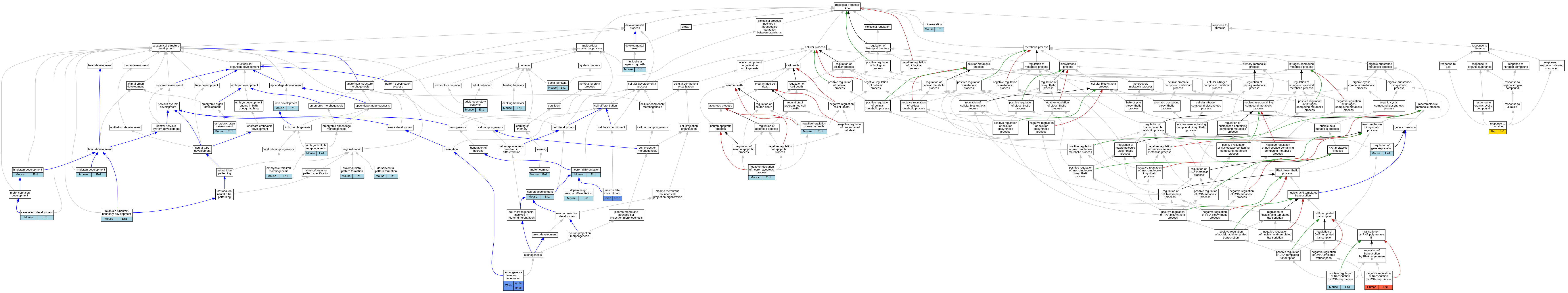
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0001227 | DNA-binding transcription repressor activity, RNA polymerase II-specific | EN1 | IDA | | Human | PMID:21672318 |
| Molecular Function | GO:0005515 | protein binding | En1 | IPI | PR:Q8VIJ6 | Mouse | PMID:23863478 |
| Molecular Function | GO:0000978 | RNA polymerase II cis-regulatory region sequence-specific DNA binding | EN1 | IDA | | Human | PMID:21672318 |
| Molecular Function | GO:1990837 | sequence-specific double-stranded DNA binding | EN1 | IDA | | Human | PMID:28473536 |
| Cellular Component | GO:0016020 | membrane | En1 | IDA | | Rat | PMID:9169834|RGD:5687201 |
| Cellular Component | GO:0005634 | nucleus | EN1 | IDA | | Human | PMID:24399192 |
| Cellular Component | GO:0005634 | nucleus | En1 | IDA | | Rat | PMID:9169834|RGD:5687201 |
| Biological Process | GO:0008344 | adult locomotory behavior | En1 | IMP | | Mouse | MGI:MGI:3697962|PMID:17267560 |
| Biological Process | GO:0060385 | axonogenesis involved in innervation | en1a | IGI | ZFIN:ZDB-MRPHLNO-171117-11,ZFIN:ZDB-MRPHLNO-171117-7,ZFIN:ZDB-MRPHLNO-171117-9 | Zfish | PMID:28807781 |
| Biological Process | GO:0060385 | axonogenesis involved in innervation | en1a | IGI | ZFIN:ZDB-MRPHLNO-171117-10,ZFIN:ZDB-MRPHLNO-171117-11,ZFIN:ZDB-MRPHLNO-171117-6,ZFIN:ZDB-MRPHLNO-171117-7,ZFIN:ZDB-MRPHLNO-171117-8,ZFIN:ZDB-MRPHLNO-171117-9 | Zfish | PMID:28807781 |
| Biological Process | GO:0060385 | axonogenesis involved in innervation | en1b | IGI | ZFIN:ZDB-MRPHLNO-171117-10,ZFIN:ZDB-MRPHLNO-171117-11,ZFIN:ZDB-MRPHLNO-171117-6,ZFIN:ZDB-MRPHLNO-171117-7,ZFIN:ZDB-MRPHLNO-171117-8,ZFIN:ZDB-MRPHLNO-171117-9 | Zfish | PMID:28807781 |
| Biological Process | GO:0060385 | axonogenesis involved in innervation | en1b | IGI | ZFIN:ZDB-MRPHLNO-171117-11,ZFIN:ZDB-MRPHLNO-171117-7,ZFIN:ZDB-MRPHLNO-171117-9 | Zfish | PMID:28807781 |
| Biological Process | GO:0021549 | cerebellum development | En1 | IGI | UniProtKB:P04426 | Mouse | MGI:MGI:82938|PMID:8848044 |
| Biological Process | GO:0071542 | dopaminergic neuron differentiation | En1 | IGI | MGI:MGI:95390 | Mouse | PMID:15175251 |
| Biological Process | GO:0071542 | dopaminergic neuron differentiation | En1 | IGI | MGI:MGI:95390 | Mouse | PMID:15340832 |
| Biological Process | GO:0009953 | dorsal/ventral pattern formation | En1 | IMP | MGI:MGI:1857163 | Mouse | PMID:9362463 |
| Biological Process | GO:0009953 | dorsal/ventral pattern formation | En1 | IMP | MGI:MGI:1857163 | Mouse | PMID:8684466 |
| Biological Process | GO:0042756 | drinking behavior | En1 | IMP | | Mouse | MGI:MGI:3697962|PMID:17267560 |
| Biological Process | GO:1990403 | embryonic brain development | En1 | IMP | | Mouse | MGI:MGI:75252|PMID:7624797 |
| Biological Process | GO:0035115 | embryonic forelimb morphogenesis | En1 | IGI | MGI:MGI:98961 | Mouse | PMID:9362463 |
| Biological Process | GO:0030326 | embryonic limb morphogenesis | En1 | IMP | | Mouse | MGI:MGI:75252|PMID:7624797 |
| Biological Process | GO:0030326 | embryonic limb morphogenesis | En1 | IGI | MGI:MGI:1329040 | Mouse | PMID:15242796 |
| Biological Process | GO:0030326 | embryonic limb morphogenesis | En1 | IMP | MGI:MGI:1857163 | Mouse | PMID:8684466 |
| Biological Process | GO:0030326 | embryonic limb morphogenesis | En1 | IGI | MGI:MGI:98961 | Mouse | PMID:15242796 |
| Biological Process | GO:0030902 | hindbrain development | En1 | IGI | MGI:MGI:95390 | Mouse | PMID:15607945 |
| Biological Process | GO:0060173 | limb development | En1 | IMP | MGI:MGI:1857163 | Mouse | PMID:23863478 |
| Biological Process | GO:0030901 | midbrain development | En1 | IGI | UniProtKB:P04426 | Mouse | MGI:MGI:82938|PMID:8848044 |
| Biological Process | GO:0030901 | midbrain development | En1 | IGI | MGI:MGI:95390 | Mouse | PMID:15607945 |
| Biological Process | GO:0030901 | midbrain development | En1 | IGI | MGI:MGI:95390 | Mouse | PMID:15175251 |
| Biological Process | GO:0030917 | midbrain-hindbrain boundary development | En1 | IMP | MGI:MGI:1857163 | Mouse | PMID:15607945 |
| Biological Process | GO:0061743 | motor learning | En1 | IMP | | Mouse | MGI:MGI:3697962|PMID:17267560 |
| Biological Process | GO:0035264 | multicellular organism growth | En1 | IMP | MGI:MGI:1857163 | Mouse | PMID:23863478 |
| Biological Process | GO:0043524 | negative regulation of neuron apoptotic process | En1 | IGI | UniProtKB:P09066 | Mouse | MGI:MGI:3578062|PMID:15175251 |
| Biological Process | GO:1901215 | negative regulation of neuron death | En1 | IMP | | Mouse | MGI:MGI:3697962|PMID:17267560 |
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | EN1 | IDA | | Human | PMID:21672318 |
| Biological Process | GO:0048666 | neuron development | En1 | IGI | MGI:MGI:95390 | Mouse | PMID:11312297 |
| Biological Process | GO:0048666 | neuron development | En1 | IMP | MGI:MGI:1857163 | Mouse | PMID:23863478 |
| Biological Process | GO:0030182 | neuron differentiation | En1 | IGI | MGI:MGI:95390 | Mouse | PMID:15340832 |
| Biological Process | GO:0030182 | neuron differentiation | En1 | IGI | MGI:MGI:95390 | Mouse | PMID:15607945 |
| Biological Process | GO:0048663 | neuron fate commitment | en1b | IEP | | Zfish | PMID:15215305 |
| Biological Process | GO:0043473 | pigmentation | En1 | IMP | MGI:MGI:1857163 | Mouse | PMID:9362463 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | En1 | IDA | | Mouse | PMID:9102311 |
| Biological Process | GO:0009954 | proximal/distal pattern formation | En1 | IMP | MGI:MGI:1857163 | Mouse | PMID:8684466 |
| Biological Process | GO:0009954 | proximal/distal pattern formation | En1 | IMP | MGI:MGI:1857163 | Mouse | PMID:9362463 |
| Biological Process | GO:0010468 | regulation of gene expression | En1 | IMP | MGI:MGI:1857163 | Mouse | PMID:23863478 |
| Biological Process | GO:0010468 | regulation of gene expression | En1 | IGI | MGI:MGI:1100498 | Mouse | PMID:23863478 |
| Biological Process | GO:0042220 | response to cocaine | En1 | IEP | | Rat | PMID:17070804|RGD:5687200 |
| Biological Process | GO:0035176 | social behavior | En1 | IMP | | Mouse | MGI:MGI:3697962|PMID:17267560 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
 Analysis Tools
Analysis Tools