|
Source |
Alliance of Genome Resources |
| Category | ID | Classification term | Gene | Evidence | Inferred from | Organism | Reference |
|---|---|---|---|---|---|---|---|
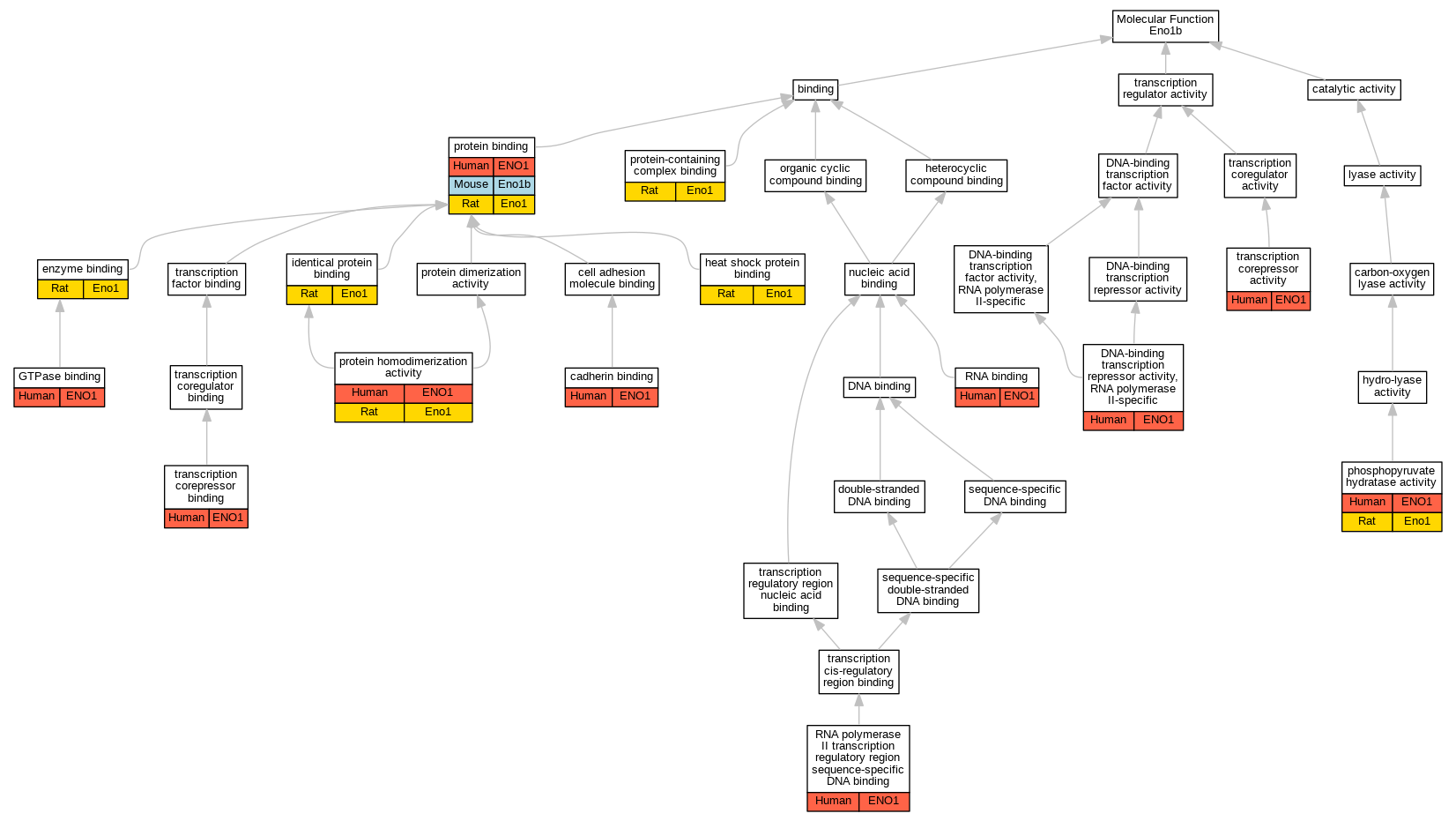
| Molecular Function | GO:0045296 | cadherin binding | ENO1 | HDA | Human | PMID:25468996 | |
| Molecular Function | GO:0001227 | DNA-binding transcription repressor activity, RNA polymerase II-specific | ENO1 | IDA | Human | PMID:2005901 | |
| Molecular Function | GO:0019899 | enzyme binding | Eno1 | IPI | UniProtKB:P07323 | Rat | PMID:14499952|RGD:12793007 |
| Molecular Function | GO:0019899 | enzyme binding | Eno1 | IPI | UniProtKB:P07323 | Rat | PMID:15041191|RGD:2302788 |
| Molecular Function | GO:0051020 | GTPase binding | ENO1 | IPI | UniProtKB:P32455 | Human | PMID:24337748 |
| Molecular Function | GO:0031072 | heat shock protein binding | Eno1 | IPI | RGD:621725 | Rat | PMID:21958194|RGD:10059326 |
| Molecular Function | GO:0042802 | identical protein binding | Eno1 | IPI | RGD:2553 | Rat | PMID:7086434|RGD:2302787 |
| Molecular Function | GO:0004634 | phosphopyruvate hydratase activity | ENO1 | IDA | Human | PMID:29775581 | |
| Molecular Function | GO:0004634 | phosphopyruvate hydratase activity | ENO1 | IMP | Human | PMID:3529090 | |
| Molecular Function | GO:0004634 | phosphopyruvate hydratase activity | Eno1 | IDA | Rat | PMID:7964722|RGD:12792993 | |
| Molecular Function | GO:0005515 | protein binding | ENO1 | IPI | UniProtKB:P00747 | Human | PMID:12666133 |
| Molecular Function | GO:0005515 | protein binding | ENO1 | IPI | UniProtKB:P63104 | Human | PMID:15161933 |
| Molecular Function | GO:0005515 | protein binding | ENO1 | IPI | UniProtKB:P42858 | Human | PMID:17500595 |
| Molecular Function | GO:0005515 | protein binding | ENO1 | IPI | UniProtKB:P22303 | Human | PMID:18194455 |
| Molecular Function | GO:0005515 | protein binding | ENO1 | IPI | UniProtKB:P00747 | Human | PMID:19433310 |
| Molecular Function | GO:0005515 | protein binding | ENO1 | IPI | UniProtKB:P12004 | Human | PMID:20849852 |
| Molecular Function | GO:0005515 | protein binding | ENO1 | IPI | UniProtKB:O75952-1 | Human | PMID:21274509 |
| Molecular Function | GO:0005515 | protein binding | ENO1 | IPI | UniProtKB:P21754 | Human | PMID:23355646 |
| Molecular Function | GO:0005515 | protein binding | ENO1 | IPI | UniProtKB:Q05996 | Human | PMID:23355646 |
| Molecular Function | GO:0005515 | protein binding | ENO1 | IPI | UniProtKB:Q12836 | Human | PMID:23355646 |
| Molecular Function | GO:0005515 | protein binding | ENO1 | IPI | UniProtKB:P63104 | Human | PMID:23414517 |
| Molecular Function | GO:0005515 | protein binding | ENO1 | IPI | UniProtKB:Q8WZ42 | Human | PMID:23414517 |
| Molecular Function | GO:0005515 | protein binding | ENO1 | IPI | UniProtKB:Q9NX76 | Human | PMID:28813417 |
| Molecular Function | GO:0005515 | protein binding | ENO1 | IPI | UniProtKB:P42858 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | Eno1b | IPI | PR:Q8C042 | Mouse | PMID:23446454 |
| Molecular Function | GO:0005515 | protein binding | Eno1b | IPI | PR:O70250 | Mouse | PMID:23446454 |
| Molecular Function | GO:0005515 | protein binding | Eno1 | IPI | UniProtKB:Q5VU43-11 | Rat | PMID:21569246|RGD:10047194 |
| Molecular Function | GO:0005515 | protein binding | Eno1 | IPI | UniProtKB:Q01177 | Rat | PMID:7964722|RGD:12792993 |
| Molecular Function | GO:0042803 | protein homodimerization activity | ENO1 | IDA | Human | PMID:3529090 | |
| Molecular Function | GO:0042803 | protein homodimerization activity | Eno1 | IDA | Rat | PMID:7964722|RGD:12792993 | |
| Molecular Function | GO:0044877 | protein-containing complex binding | Eno1 | IDA | Rat | PMID:15041191|RGD:2302788 | |
| Molecular Function | GO:0044877 | protein-containing complex binding | Eno1 | IDA | Rat | PMID:7086434|RGD:2302787 | |
| Molecular Function | GO:0003723 | RNA binding | ENO1 | HDA | Human | PMID:22658674 | |
| Molecular Function | GO:0000977 | RNA polymerase II transcription regulatory region sequence-specific DNA binding | ENO1 | IDA | Human | PMID:2005901 | |
| Molecular Function | GO:0003714 | transcription corepressor activity | ENO1 | IDA | Human | PMID:11134351 | |
| Molecular Function | GO:0001222 | transcription corepressor binding | ENO1 | IPI | UniProtKB:P0DI82 | Human | PMID:11134351 |
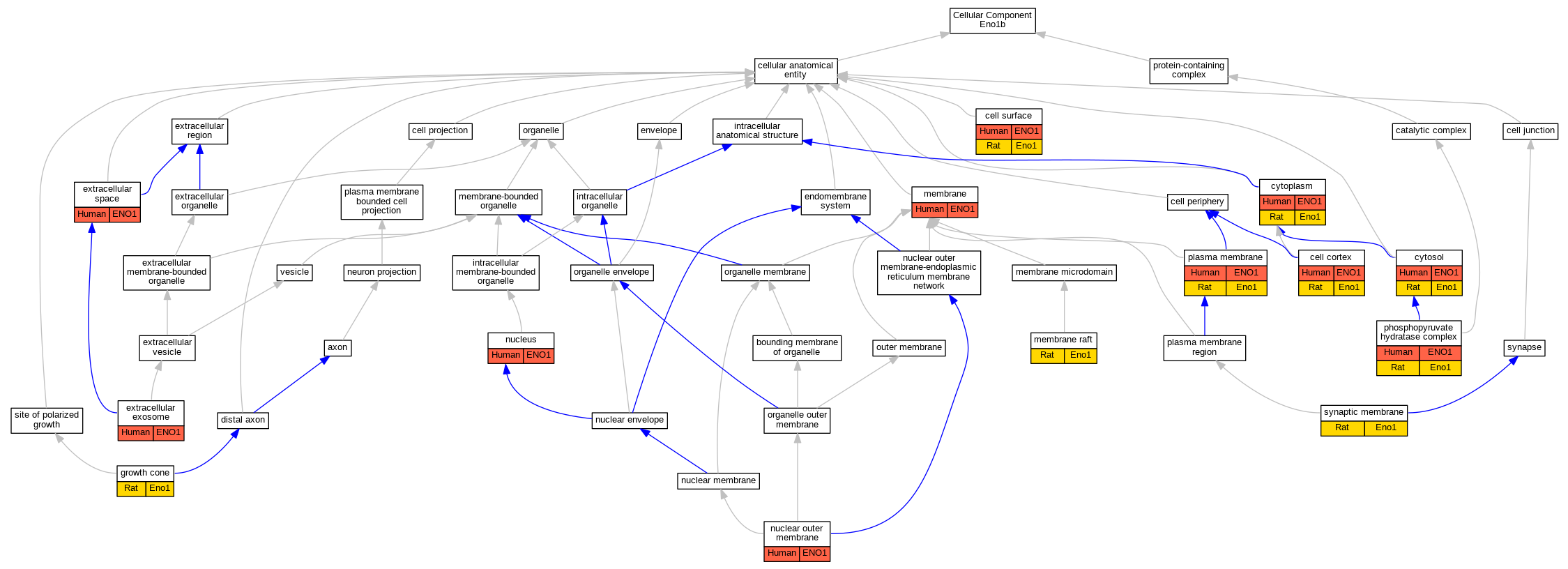
| Cellular Component | GO:0005938 | cell cortex | ENO1 | IDA | Human | PMID:19433310 | |
| Cellular Component | GO:0005938 | cell cortex | Eno1 | IDA | Rat | PMID:19433310|RGD:12792998 | |
| Cellular Component | GO:0009986 | cell surface | ENO1 | IDA | Human | PMID:12666133 | |
| Cellular Component | GO:0009986 | cell surface | ENO1 | IDA | Human | PMID:19433310 | |
| Cellular Component | GO:0009986 | cell surface | Eno1 | IDA | Rat | PMID:7964722|RGD:12792993 | |
| Cellular Component | GO:0005737 | cytoplasm | ENO1 | IDA | Human | GO_REF:0000054 | |
| Cellular Component | GO:0005737 | cytoplasm | ENO1 | IDA | Human | PMID:15459207 | |
| Cellular Component | GO:0005737 | cytoplasm | ENO1 | IDA | Human | PMID:25468996 | |
| Cellular Component | GO:0005737 | cytoplasm | Eno1 | IDA | Rat | PMID:15459207|RGD:12793039 | |
| Cellular Component | GO:0005829 | cytosol | ENO1 | IDA | Human | GO_REF:0000052 | |
| Cellular Component | GO:0005829 | cytosol | ENO1 | IDA | Human | PMID:12666133 | |
| Cellular Component | GO:0005829 | cytosol | ENO1 | IDA | Human | PMID:19433310 | |
| Cellular Component | GO:0005829 | cytosol | Eno1 | IDA | Rat | PMID:14499952|RGD:12793007 | |
| Cellular Component | GO:0070062 | extracellular exosome | ENO1 | HDA | Human | PMID:11487543 | |
| Cellular Component | GO:0070062 | extracellular exosome | ENO1 | HDA | Human | PMID:12519789 | |
| Cellular Component | GO:0070062 | extracellular exosome | ENO1 | HDA | Human | PMID:19199708 | |
| Cellular Component | GO:0070062 | extracellular exosome | ENO1 | HDA | Human | PMID:20458337 | |
| Cellular Component | GO:0070062 | extracellular exosome | ENO1 | HDA | Human | PMID:21362503 | |
| Cellular Component | GO:0070062 | extracellular exosome | ENO1 | HDA | Human | PMID:23533145 | |
| Cellular Component | GO:0005615 | extracellular space | ENO1 | HDA | Human | PMID:16502470 | |
| Cellular Component | GO:0005615 | extracellular space | ENO1 | HDA | Human | PMID:22664934 | |
| Cellular Component | GO:0005615 | extracellular space | ENO1 | HDA | Human | PMID:23580065 | |
| Cellular Component | GO:0030426 | growth cone | Eno1 | IDA | Rat | PMID:19433310|RGD:12792998 | |
| Cellular Component | GO:0016020 | membrane | ENO1 | IDA | Human | PMID:19433310 | |
| Cellular Component | GO:0016020 | membrane | ENO1 | HDA | Human | PMID:19946888 | |
| Cellular Component | GO:0045121 | membrane raft | Eno1 | IDA | Rat | PMID:14499952|RGD:12793007 | |
| Cellular Component | GO:0005640 | nuclear outer membrane | ENO1 | IDA | Human | PMID:11134351 | |
| Cellular Component | GO:0005634 | nucleus | ENO1 | HDA | Human | PMID:21630459 | |
| Cellular Component | GO:0000015 | phosphopyruvate hydratase complex | ENO1 | IDA | Human | PMID:3529090 | |
| Cellular Component | GO:0000015 | phosphopyruvate hydratase complex | Eno1 | IDA | Rat | PMID:15041191|RGD:2302788 | |
| Cellular Component | GO:0005886 | plasma membrane | ENO1 | IDA | Human | GO_REF:0000052 | |
| Cellular Component | GO:0005886 | plasma membrane | ENO1 | IDA | Human | PMID:12666133 | |
| Cellular Component | GO:0005886 | plasma membrane | Eno1 | IDA | Rat | PMID:7964722|RGD:12792993 | |
| Cellular Component | GO:0097060 | synaptic membrane | Eno1 | IDA | Rat | PMID:15041191|RGD:2302788 | |
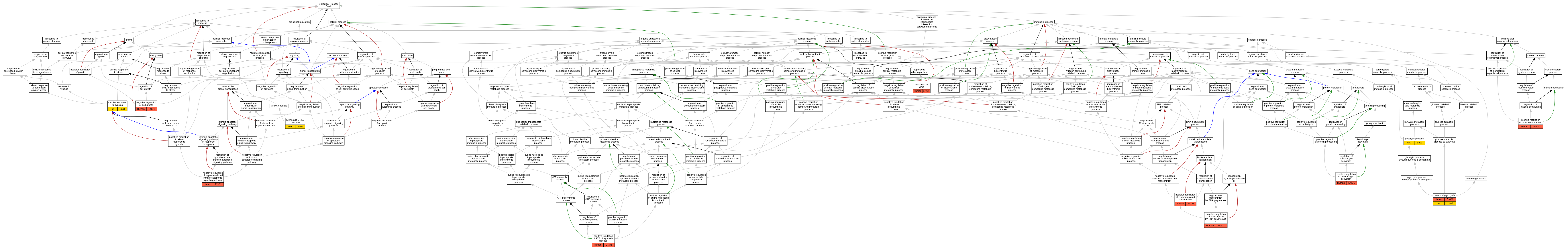
| Biological Process | GO:0061621 | canonical glycolysis | ENO1 | IDA | Human | PMID:29775581 | |
| Biological Process | GO:0061621 | canonical glycolysis | ENO1 | IMP | Human | PMID:3529090 | |
| Biological Process | GO:0061621 | canonical glycolysis | Eno1 | IDA | Rat | PMID:15041191|RGD:2302788 | |
| Biological Process | GO:0071456 | cellular response to hypoxia | Eno1 | IDA | Rat | PMID:15459207|RGD:12793039 | |
| Biological Process | GO:0070371 | ERK1 and ERK2 cascade | Eno1 | IDA | Rat | PMID:15459207|RGD:12793039 | |
| Biological Process | GO:0006096 | glycolytic process | Eno1 | IDA | Rat | PMID:7964722|RGD:12792993 | |
| Biological Process | GO:0030308 | negative regulation of cell growth | ENO1 | IDA | Human | PMID:10082554 | |
| Biological Process | GO:0045892 | negative regulation of DNA-templated transcription | ENO1 | IDA | Human | PMID:10082554 | |
| Biological Process | GO:0045892 | negative regulation of DNA-templated transcription | ENO1 | IMP | Human | PMID:10681589 | |
| Biological Process | GO:0045892 | negative regulation of DNA-templated transcription | ENO1 | IDA | Human | PMID:11134351 | |
| Biological Process | GO:1903298 | negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway | ENO1 | IDA | Human | PMID:15459207 | |
| Biological Process | GO:1903298 | negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway | ENO1 | IGI | UniProtKB:Q01986 | Human | PMID:15459207 |
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | ENO1 | IDA | Human | PMID:2005901 | |
| Biological Process | GO:2001171 | positive regulation of ATP biosynthetic process | ENO1 | IDA | Human | PMID:15459207 | |
| Biological Process | GO:2001171 | positive regulation of ATP biosynthetic process | ENO1 | IGI | UniProtKB:Q01986 | Human | PMID:15459207 |
| Biological Process | GO:0045933 | positive regulation of muscle contraction | ENO1 | IGI | UniProtKB:Q01986 | Human | PMID:15459207 |
| Biological Process | GO:0010756 | positive regulation of plasminogen activation | ENO1 | IMP | Human | PMID:12666133 | |
| Biological Process | GO:0009615 | response to virus | ENO1 | IEP | Human | PMID:16548883 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 06/12/2024 MGI 6.13 |

|
|
|
||