| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
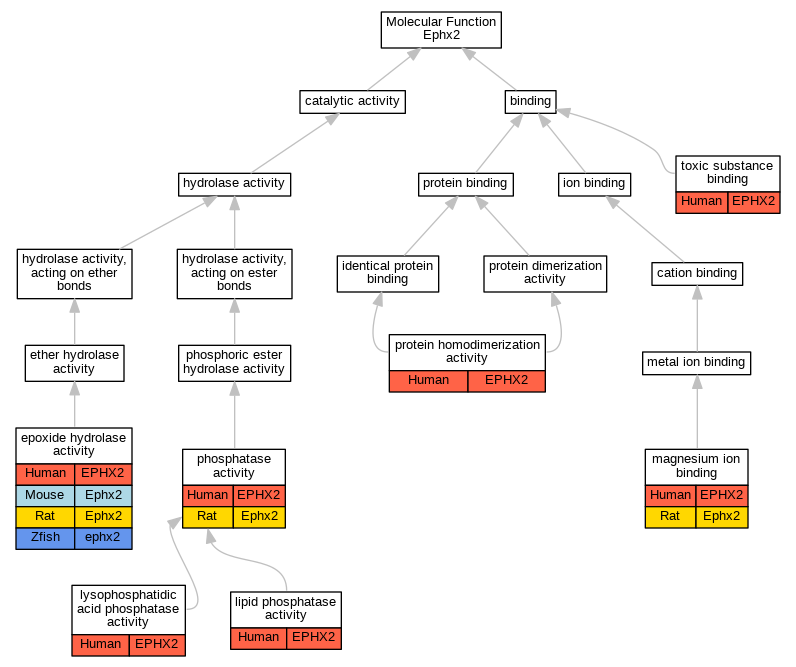
| Molecular Function | GO:0004301 | epoxide hydrolase activity | EPHX2 | IDA | | Human | PMID:10862610 |
| Molecular Function | GO:0004301 | epoxide hydrolase activity | EPHX2 | IDA | | Human | PMID:21217101 |
| Molecular Function | GO:0004301 | epoxide hydrolase activity | EPHX2 | IDA | | Human | PMID:22798687 |
| Molecular Function | GO:0004301 | epoxide hydrolase activity | EPHX2 | IDA | | Human | PMID:8342951 |
| Molecular Function | GO:0004301 | epoxide hydrolase activity | Ephx2 | IMP | | Mouse | MGI:MGI:4947022|PMID:21217101 |
| Molecular Function | GO:0004301 | epoxide hydrolase activity | Ephx2 | IDA | | Rat | PMID:21217101|RGD:21201254 |
| Molecular Function | GO:0004301 | epoxide hydrolase activity | Ephx2 | IDA | | Rat | PMID:8349641|RGD:728835 |
| Molecular Function | GO:0004301 | epoxide hydrolase activity | ephx2 | IDA | | Zfish | PMID:22665795 |
| Molecular Function | GO:0042577 | lipid phosphatase activity | EPHX2 | IDA | | Human | PMID:12574510 |
| Molecular Function | GO:0052642 | lysophosphatidic acid phosphatase activity | EPHX2 | IDA | | Human | PMID:22217705 |
| Molecular Function | GO:0052642 | lysophosphatidic acid phosphatase activity | EPHX2 | IDA | | Human | PMID:22387545 |
| Molecular Function | GO:0000287 | magnesium ion binding | EPHX2 | IDA | | Human | PMID:12574510 |
| Molecular Function | GO:0000287 | magnesium ion binding | Ephx2 | IMP | | Rat | PMID:12574508|RGD:1581955 |
| Molecular Function | GO:0016791 | phosphatase activity | EPHX2 | IDA | | Human | PMID:12574508 |
| Molecular Function | GO:0016791 | phosphatase activity | EPHX2 | IDA | | Human | PMID:15196990 |
| Molecular Function | GO:0016791 | phosphatase activity | Ephx2 | IDA | | Rat | PMID:12574508|RGD:1581955 |
| Molecular Function | GO:0042803 | protein homodimerization activity | EPHX2 | IDA | | Human | PMID:15096040 |
| Molecular Function | GO:0042803 | protein homodimerization activity | EPHX2 | IDA | | Human | PMID:18513744 |
| Molecular Function | GO:0015643 | toxic substance binding | EPHX2 | IDA | | Human | PMID:15096040 |
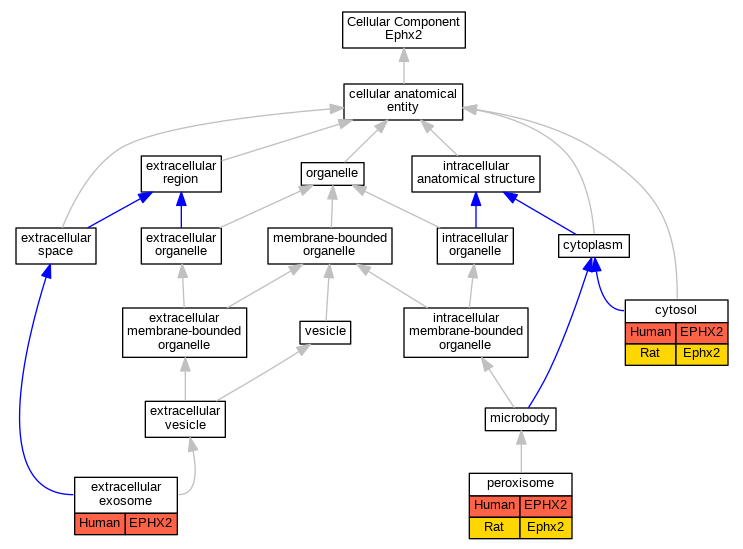
| Cellular Component | GO:0005829 | cytosol | EPHX2 | IDA | | Human | GO_REF:0000052 |
| Cellular Component | GO:0005829 | cytosol | EPHX2 | IDA | | Human | PMID:16314446 |
| Cellular Component | GO:0005829 | cytosol | EPHX2 | IDA | | Human | PMID:18513744 |
| Cellular Component | GO:0005829 | cytosol | EPHX2 | IDA | | Human | PMID:8342951 |
| Cellular Component | GO:0005829 | cytosol | Ephx2 | IDA | | Rat | PMID:10445750|RGD:1581957 |
| Cellular Component | GO:0070062 | extracellular exosome | EPHX2 | HDA | | Human | PMID:19056867 |
| Cellular Component | GO:0070062 | extracellular exosome | EPHX2 | HDA | | Human | PMID:23533145 |
| Cellular Component | GO:0005777 | peroxisome | EPHX2 | IDA | | Human | PMID:16314446 |
| Cellular Component | GO:0005777 | peroxisome | EPHX2 | IDA | | Human | PMID:18513744 |
| Cellular Component | GO:0005777 | peroxisome | Ephx2 | IDA | | Rat | PMID:3181162|RGD:1581960 |
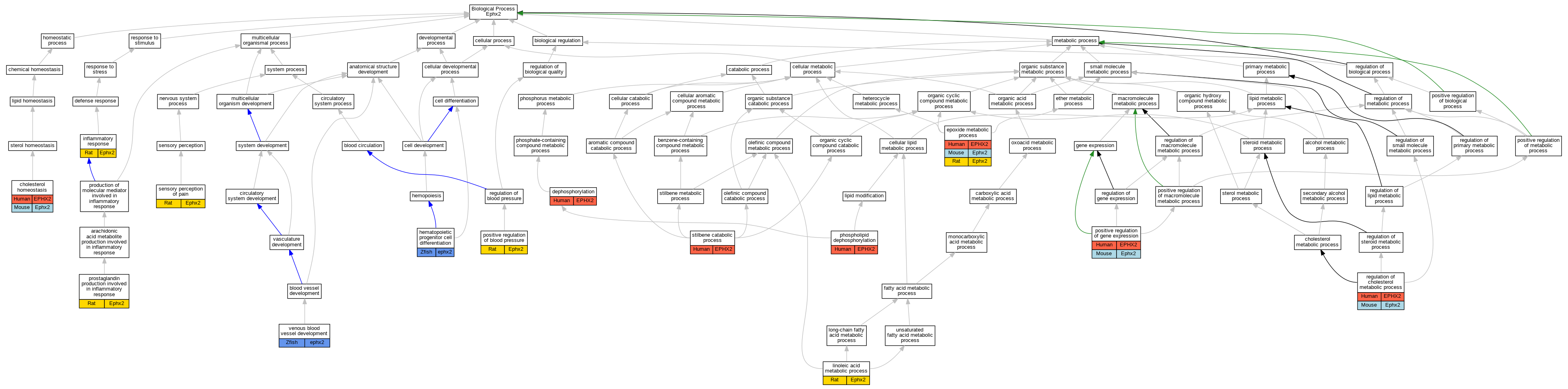
| Biological Process | GO:0042632 | cholesterol homeostasis | EPHX2 | IDA | | Human | PMID:18974052 |
| Biological Process | GO:0042632 | cholesterol homeostasis | Ephx2 | IDA | | Mouse | MGI:MGI:3829349|PMID:18974052 |
| Biological Process | GO:0016311 | dephosphorylation | EPHX2 | IDA | | Human | PMID:12574508 |
| Biological Process | GO:0016311 | dephosphorylation | EPHX2 | IDA | | Human | PMID:22217705 |
| Biological Process | GO:0097176 | epoxide metabolic process | EPHX2 | IDA | | Human | PMID:21217101 |
| Biological Process | GO:0097176 | epoxide metabolic process | EPHX2 | IDA | | Human | PMID:22798687 |
| Biological Process | GO:0097176 | epoxide metabolic process | Ephx2 | IMP | | Mouse | MGI:MGI:4947022|PMID:21217101 |
| Biological Process | GO:0097176 | epoxide metabolic process | Ephx2 | IDA | | Rat | PMID:21217101|RGD:21201254 |
| Biological Process | GO:0002244 | hematopoietic progenitor cell differentiation | ephx2 | IMP | ZFIN:ZDB-MRPHLNO-120718-1 | Zfish | PMID:22665795 |
| Biological Process | GO:0006954 | inflammatory response | Ephx2 | IMP | | Rat | PMID:15684051|RGD:1581952 |
| Biological Process | GO:0043651 | linoleic acid metabolic process | Ephx2 | IMP | | Rat | PMID:16962614|RGD:5688387 |
| Biological Process | GO:0046839 | phospholipid dephosphorylation | EPHX2 | IDA | | Human | PMID:12574510 |
| Biological Process | GO:0045777 | positive regulation of blood pressure | Ephx2 | IMP | | Rat | PMID:11882632|RGD:728682 |
| Biological Process | GO:0010628 | positive regulation of gene expression | EPHX2 | IDA | | Human | PMID:18974052 |
| Biological Process | GO:0010628 | positive regulation of gene expression | Ephx2 | IDA | | Mouse | MGI:MGI:3829349|PMID:18974052 |
| Biological Process | GO:0002539 | prostaglandin production involved in inflammatory response | Ephx2 | IMP | | Rat | PMID:16962614|RGD:5688387 |
| Biological Process | GO:0090181 | regulation of cholesterol metabolic process | EPHX2 | IMP | | Human | PMID:18974052 |
| Biological Process | GO:0090181 | regulation of cholesterol metabolic process | Ephx2 | IMP | | Mouse | MGI:MGI:3829349|PMID:18974052 |
| Biological Process | GO:0019233 | sensory perception of pain | Ephx2 | IMP | | Rat | PMID:16962614|RGD:5688387 |
| Biological Process | GO:0046272 | stilbene catabolic process | EPHX2 | IDA | | Human | PMID:8342951 |
| Biological Process | GO:0060841 | venous blood vessel development | ephx2 | IMP | ZFIN:ZDB-MRPHLNO-120718-1 | Zfish | PMID:22665795 |
| Biological Process | GO:0060841 | venous blood vessel development | ephx2 | IMP | ZFIN:ZDB-MRPHLNO-120718-2 | Zfish | PMID:22665795 |
 Analysis Tools
Analysis Tools