| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
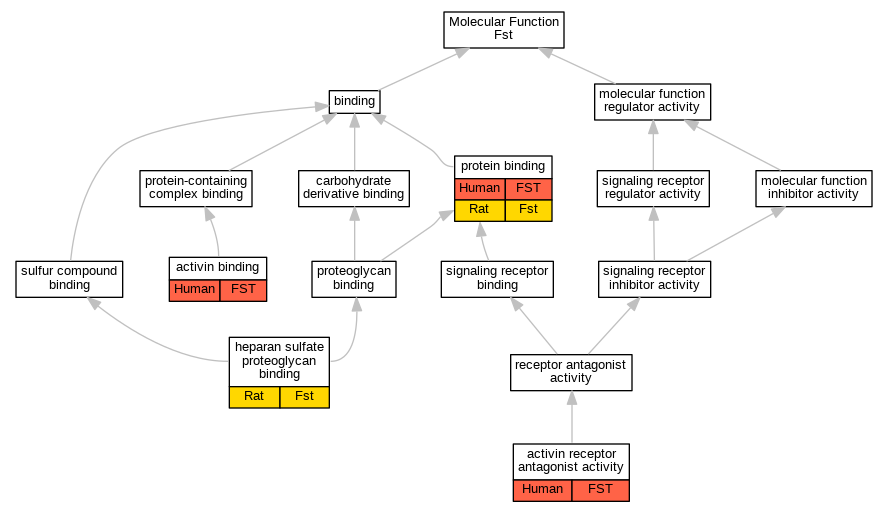
| Molecular Function | GO:0048185 | activin binding | FST | IPI | UniProtKB:P08476 | Human | PMID:12697670 |
| Molecular Function | GO:0048185 | activin binding | FST | IPI | UniProtKB:P09529 | Human | PMID:12697670 |
| Molecular Function | GO:0038102 | activin receptor antagonist activity | FST | IMP | | Human | PMID:12702211 |
| Molecular Function | GO:0043395 | heparan sulfate proteoglycan binding | Fst | IDA | | Rat | PMID:12867435|RGD:1601260 |
| Molecular Function | GO:0005515 | protein binding | FST | IPI | UniProtKB:P08476 | Human | PMID:16198295 |
| Molecular Function | GO:0005515 | protein binding | FST | IPI | UniProtKB:P03950 | Human | PMID:17991437 |
| Molecular Function | GO:0005515 | protein binding | FST | IPI | UniProtKB:Q14689 | Human | PMID:20860622 |
| Molecular Function | GO:0005515 | protein binding | FST | IPI | UniProtKB:P10599 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | FST | IPI | UniProtKB:P49639 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | FST | IPI | UniProtKB:Q02930-3 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | FST | IPI | UniProtKB:Q8NEC5 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | FST | IPI | UniProtKB:Q8TAU3 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | FST | IPI | UniProtKB:Q96SQ5 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | FST | IPI | UniProtKB:Q9BXY8 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | Fst | IPI | UniProtKB:P08476 | Rat | PMID:16482217|RGD:8554128 |
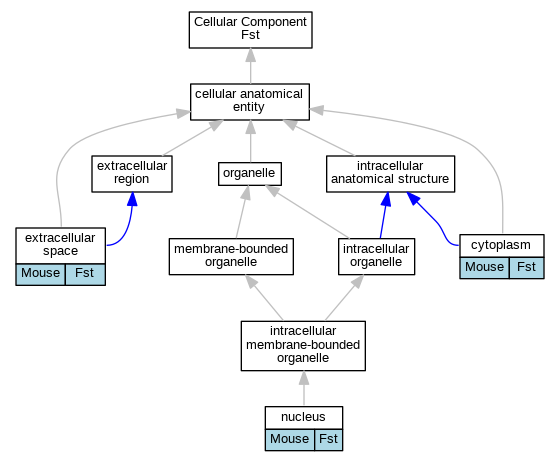
| Cellular Component | GO:0005737 | cytoplasm | Fst | IDA | | Mouse | PMID:20032047 |
| Cellular Component | GO:0005615 | extracellular space | Fst | HDA | | Mouse | MGI:MGI:5640941|PMID:24006456 |
| Cellular Component | GO:0005634 | nucleus | Fst | IDA | | Mouse | PMID:20032047 |
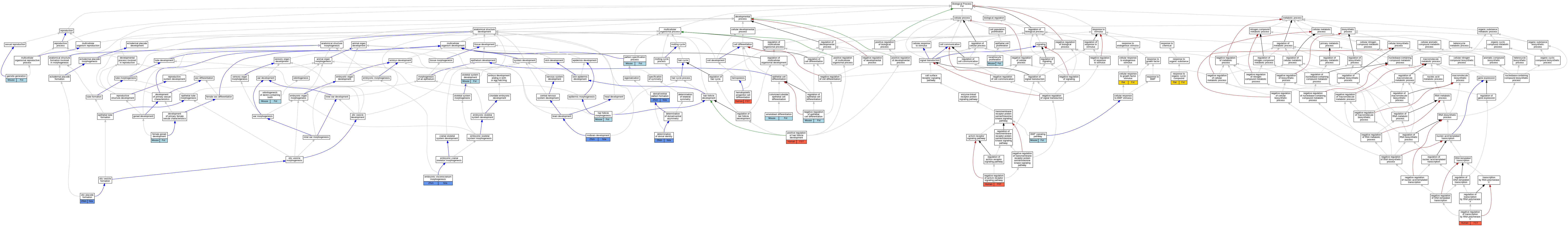
| Biological Process | GO:0036305 | ameloblast differentiation | Fst | IMP | MGI:MGI:1857467 | Mouse | PMID:15525533 |
| Biological Process | GO:0036305 | ameloblast differentiation | Fst | IGI | MGI:MGI:88180 | Mouse | PMID:15525533 |
| Biological Process | GO:0030509 | BMP signaling pathway | Fst | IGI | MGI:MGI:88180 | Mouse | PMID:15525533 |
| Biological Process | GO:0071363 | cellular response to growth factor stimulus | Fst | IEP | | Rat | PMID:23567549|RGD:151893499 |
| Biological Process | GO:0048263 | determination of dorsal identity | fsta | IMP | | Zfish | PMID:9882485 |
| Biological Process | GO:0009953 | dorsal/ventral pattern formation | fsta | IDA | | Zfish | PMID:16890217 |
| Biological Process | GO:0009953 | dorsal/ventral pattern formation | fsta | IMP | | Zfish | PMID:9882485 |
| Biological Process | GO:0048703 | embryonic viscerocranium morphogenesis | fsta | IMP | ZFIN:ZDB-MRPHLNO-080212-3 | Zfish | PMID:23209659 |
| Biological Process | GO:0008585 | female gonad development | Fst | IMP | MGI:MGI:1857467 | Mouse | PMID:15162500 |
| Biological Process | GO:0007276 | gamete generation | Fst | IGI | MGI:MGI:98957 | Mouse | PMID:15162500 |
| Biological Process | GO:0031069 | hair follicle morphogenesis | Fst | IMP | MGI:MGI:1857467 | Mouse | PMID:12514121 |
| Biological Process | GO:0002244 | hematopoietic progenitor cell differentiation | FST | IDA | | Human | PMID:15451575 |
| Biological Process | GO:0043616 | keratinocyte proliferation | Fst | IMP | MGI:MGI:1857467 | Mouse | PMID:12514121 |
| Biological Process | GO:0030901 | midbrain development | fsta | IMP | ZFIN:ZDB-MRPHLNO-100426-5 | Zfish | PMID:20194436 |
| Biological Process | GO:0030901 | midbrain development | fsta | IMP | ZFIN:ZDB-MRPHLNO-100426-6 | Zfish | PMID:20194436 |
| Biological Process | GO:0032926 | negative regulation of activin receptor signaling pathway | FST | IDA | | Human | PMID:11948405 |
| Biological Process | GO:0032926 | negative regulation of activin receptor signaling pathway | FST | IDA | | Human | PMID:12697670 |
| Biological Process | GO:0032926 | negative regulation of activin receptor signaling pathway | FST | IMP | | Human | PMID:12702211 |
| Biological Process | GO:0030857 | negative regulation of epithelial cell differentiation | Fst | IMP | MGI:MGI:1857467 | Mouse | PMID:15525533 |
| Biological Process | GO:0030857 | negative regulation of epithelial cell differentiation | Fst | IGI | MGI:MGI:88180 | Mouse | PMID:15525533 |
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | FST | IDA | | Human | PMID:12702211 |
| Biological Process | GO:0042475 | odontogenesis of dentin-containing tooth | Fst | IMP | MGI:MGI:1857467 | Mouse | PMID:15305290 |
| Biological Process | GO:0042475 | odontogenesis of dentin-containing tooth | Fst | IMP | MGI:MGI:1857467 | Mouse | PMID:15525533 |
| Biological Process | GO:0043049 | otic placode formation | fsta | IGI | ZFIN:ZDB-MRPHLNO-050221-6,ZFIN:ZDB-MRPHLNO-060320-5,ZFIN:ZDB-MRPHLNO-101102-5 | Zfish | PMID:19418450 |
| Biological Process | GO:0007389 | pattern specification process | Fst | IMP | MGI:MGI:1857467 | Mouse | PMID:15525533 |
| Biological Process | GO:0051798 | positive regulation of hair follicle development | FST | IDA | | Human | PMID:12514121 |
| Biological Process | GO:0014070 | response to organic cyclic compound | Fst | IEP | | Rat | PMID:30599193|RGD:151893501 |
| Biological Process | GO:0001501 | skeletal system development | Fst | IGI | MGI:MGI:2670967 | Mouse | PMID:24019467 |
 Analysis Tools
Analysis Tools