|
Source |
Alliance of Genome Resources |

| Category | ID | Classification term | Gene | Evidence | Inferred from | Organism | Reference |
|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0017083 | 4-galactosyl-N-acetylglucosaminide 3-alpha-L-fucosyltransferase activity | FUT9 | IMP | Human | PMID:10622713 | |
| Molecular Function | GO:0017083 | 4-galactosyl-N-acetylglucosaminide 3-alpha-L-fucosyltransferase activity | FUT9 | IDA | Human | PMID:11278338 | |
| Molecular Function | GO:0017083 | 4-galactosyl-N-acetylglucosaminide 3-alpha-L-fucosyltransferase activity | FUT9 | IMP | Human | PMID:12107078 | |
| Molecular Function | GO:0017083 | 4-galactosyl-N-acetylglucosaminide 3-alpha-L-fucosyltransferase activity | FUT9 | IDA | Human | PMID:18395013 | |
| Molecular Function | GO:0017083 | 4-galactosyl-N-acetylglucosaminide 3-alpha-L-fucosyltransferase activity | FUT9 | IMP | Human | PMID:23192350 | |
| Molecular Function | GO:0017083 | 4-galactosyl-N-acetylglucosaminide 3-alpha-L-fucosyltransferase activity | FUT9 | IDA | Human | PMID:29593094 | |
| Molecular Function | GO:0017083 | 4-galactosyl-N-acetylglucosaminide 3-alpha-L-fucosyltransferase activity | Fut9 | IDA | Mouse | MGI:MGI:6383286|PMID:12626397 | |
| Molecular Function | GO:0046920 | alpha-(1->3)-fucosyltransferase activity | Fut9 | IMP | MGI:MGI:3044951 | Mouse | PMID:23986452 |
| Molecular Function | GO:0046920 | alpha-(1->3)-fucosyltransferase activity | Fut9 | IDA | Mouse | PMID:15364956 | |
| Molecular Function | GO:0005515 | protein binding | FUT9 | IPI | UniProtKB:O75031 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | FUT9 | IPI | UniProtKB:P58418 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | FUT9 | IPI | UniProtKB:Q8NBJ4 | Human | PMID:32296183 |
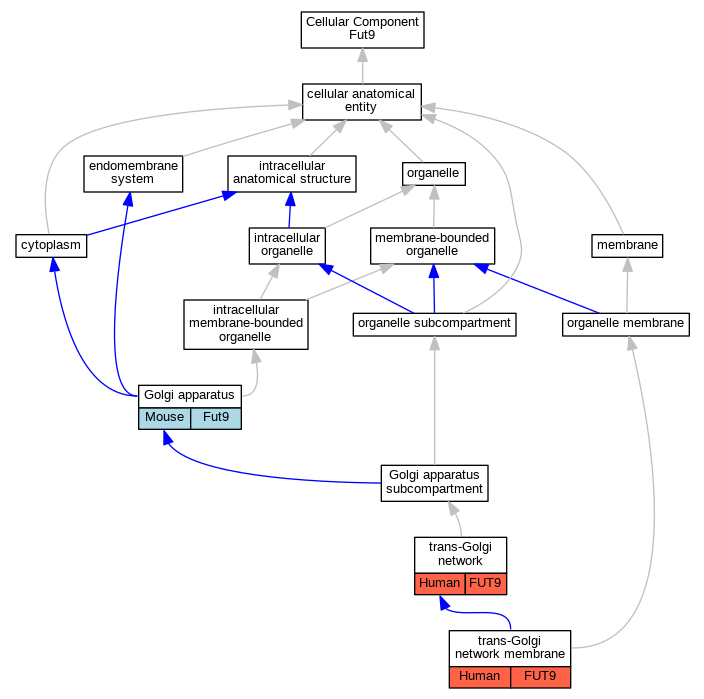
| Cellular Component | GO:0005794 | Golgi apparatus | Fut9 | IDA | Mouse | MGI:MGI:6383286|PMID:12626397 | |
| Cellular Component | GO:0005802 | trans-Golgi network | FUT9 | IDA | Human | PMID:23192350 | |
| Cellular Component | GO:0032588 | trans-Golgi network membrane | FUT9 | IDA | Human | PMID:18395013 | |
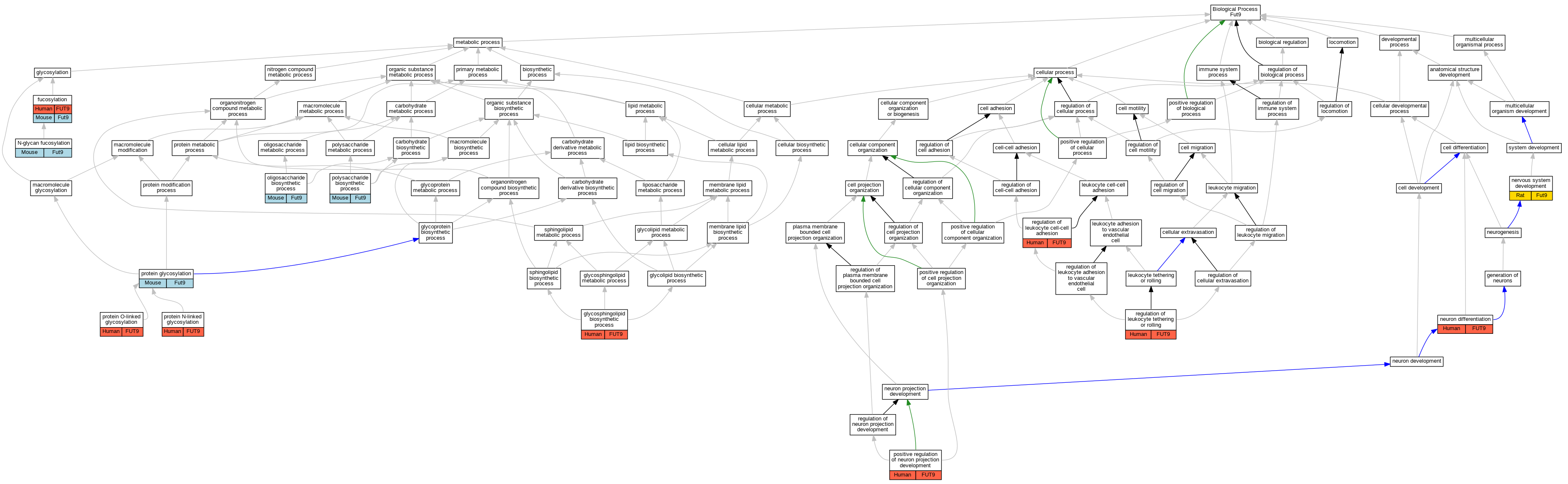
| Biological Process | GO:0036065 | fucosylation | FUT9 | IDA | Human | PMID:18395013 | |
| Biological Process | GO:0036065 | fucosylation | Fut9 | IDA | Mouse | MGI:MGI:6383286|PMID:12626397 | |
| Biological Process | GO:0006688 | glycosphingolipid biosynthetic process | FUT9 | IDA | Human | PMID:29593094 | |
| Biological Process | GO:0036071 | N-glycan fucosylation | Fut9 | IDA | Mouse | PMID:23986452 | |
| Biological Process | GO:0007399 | nervous system development | Fut9 | IDA | Rat | PMID:11675393|RGD:632768 | |
| Biological Process | GO:0030182 | neuron differentiation | FUT9 | IDA | Human | PMID:17335083 | |
| Biological Process | GO:0009312 | oligosaccharide biosynthetic process | Fut9 | IMP | MGI:MGI:3044951 | Mouse | PMID:23986452 |
| Biological Process | GO:0000271 | polysaccharide biosynthetic process | Fut9 | IMP | Mouse | MGI:MGI:3041940|PMID:15121843 | |
| Biological Process | GO:0010976 | positive regulation of neuron projection development | FUT9 | IMP | Human | PMID:23000574 | |
| Biological Process | GO:0006486 | protein glycosylation | Fut9 | IDA | Mouse | PMID:15364956 | |
| Biological Process | GO:0006487 | protein N-linked glycosylation | FUT9 | IDA | Human | PMID:29593094 | |
| Biological Process | GO:0006493 | protein O-linked glycosylation | FUT9 | IDA | Human | PMID:29593094 | |
| Biological Process | GO:1903037 | regulation of leukocyte cell-cell adhesion | FUT9 | IMP | Human | PMID:23192350 | |
| Biological Process | GO:1903236 | regulation of leukocyte tethering or rolling | FUT9 | IMP | Human | PMID:23192350 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 09/03/2024 MGI 6.24 |

|
|
|
||