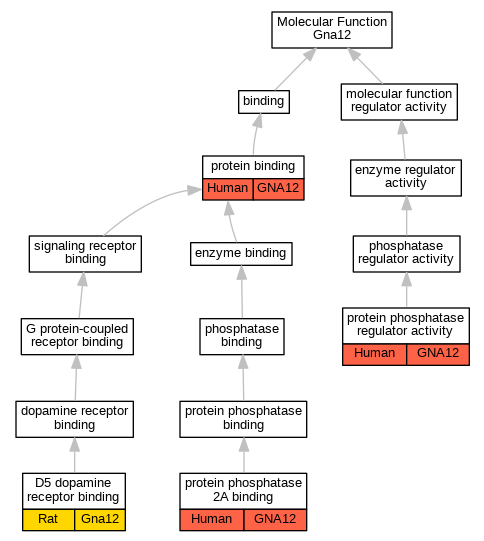
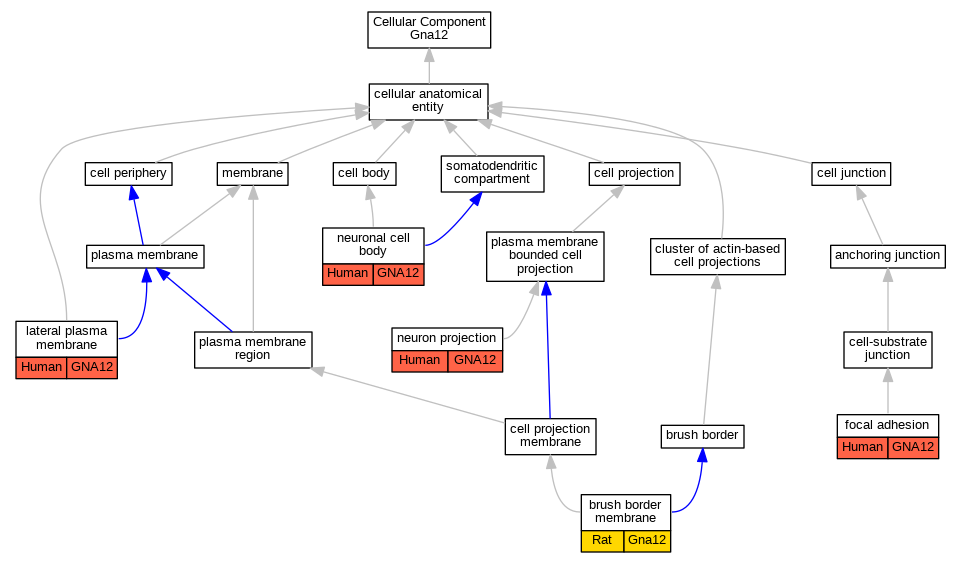
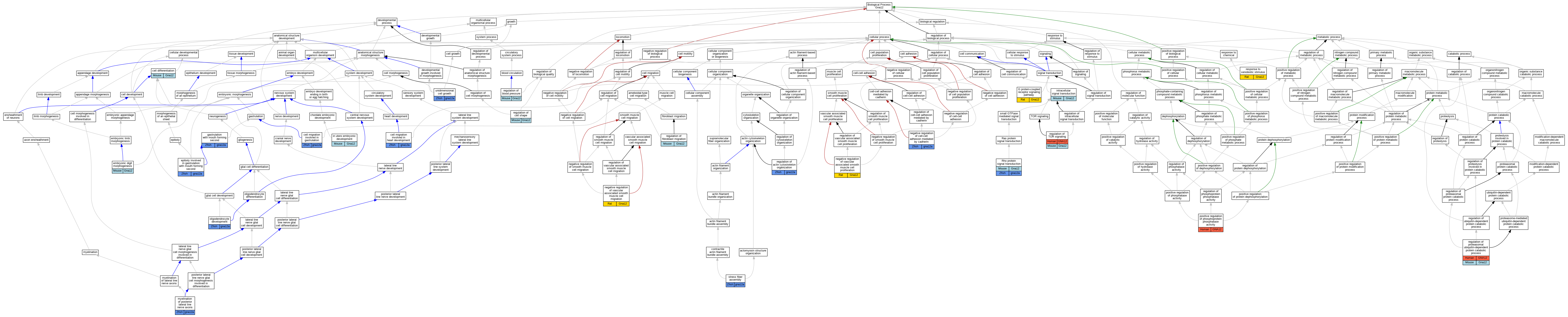
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0031752 | D5 dopamine receptor binding | Gna12 | IPI | RGD:2523 | Rat | PMID:12623966|RGD:730287 |
| Molecular Function | GO:0005515 | protein binding | GNA12 | IPI | UniProtKB:O15085 | Human | PMID:10026210 |
| Molecular Function | GO:0005515 | protein binding | GNA12 | IPI | UniProtKB:Q8NE09 | Human | PMID:18703424 |
| Molecular Function | GO:0051721 | protein phosphatase 2A binding | GNA12 | IPI | UniProtKB:P30153 | Human | PMID:15525651 |
| Molecular Function | GO:0019888 | protein phosphatase regulator activity | GNA12 | IDA | | Human | PMID:15525651 |
| Cellular Component | GO:0031526 | brush border membrane | Gna12 | IDA | | Rat | PMID:12623966|RGD:730287 |
| Cellular Component | GO:0005925 | focal adhesion | GNA12 | HDA | | Human | PMID:21423176 |
| Cellular Component | GO:0016328 | lateral plasma membrane | GNA12 | IDA | | Human | PMID:15525651 |
| Cellular Component | GO:0043005 | neuron projection | GNA12 | IDA | | Human | PMID:15525651 |
| Cellular Component | GO:0043025 | neuronal cell body | GNA12 | IDA | | Human | PMID:15525651 |
| Biological Process | GO:0030154 | cell differentiation | Gna12 | IDA | | Mouse | PMID:9305907 |
| Biological Process | GO:0042074 | cell migration involved in gastrulation | gna12a | IGI | ZFIN:ZDB-MRPHLNO-060113-1,ZFIN:ZDB-MRPHLNO-060113-3,ZFIN:ZDB-MRPHLNO-060113-4 | Zfish | PMID:15928205 |
| Biological Process | GO:0060973 | cell migration involved in heart development | gna12a | IGI | ZFIN:ZDB-GENE-030131-5024,ZFIN:ZDB-GENE-030131-8277 | Zfish | PMID:23318642 |
| Biological Process | GO:0042733 | embryonic digit morphogenesis | Gna12 | IGI | MGI:MGI:95776 | Mouse | PMID:12077299 |
| Biological Process | GO:0055113 | epiboly involved in gastrulation with mouth forming second | gna12a | IGI | ZFIN:ZDB-MRPHLNO-060113-1,ZFIN:ZDB-MRPHLNO-060113-3,ZFIN:ZDB-MRPHLNO-060113-4 | Zfish | PMID:19307601 |
| Biological Process | GO:0007186 | G protein-coupled receptor signaling pathway | Gna12 | IMP | | Rat | PMID:29453251|RGD:42721986 |
| Biological Process | GO:0001702 | gastrulation with mouth forming second | gna12a | IMP | | Zfish | PMID:15928205 |
| Biological Process | GO:0001702 | gastrulation with mouth forming second | gna12a | IGI | ZFIN:ZDB-GENO-980202-187,ZFIN:ZDB-MRPHLNO-060113-3,ZFIN:ZDB-MRPHLNO-060113-4 | Zfish | PMID:15928205 |
| Biological Process | GO:0001701 | in utero embryonic development | Gna12 | IGI | MGI:MGI:95768 | Mouse | PMID:12077299 |
| Biological Process | GO:0035556 | intracellular signal transduction | Gna12 | IDA | | Mouse | PMID:10037795 |
| Biological Process | GO:0048932 | myelination of posterior lateral line nerve axons | gna12a | IGI | ZFIN:ZDB-GENO-180713-3,ZFIN:ZDB-MRPHLNO-060113-1,ZFIN:ZDB-MRPHLNO-060113-3,ZFIN:ZDB-MRPHLNO-060113-4 | Zfish | PMID:29367382 |
| Biological Process | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin | gna12a | IGI | ZFIN:ZDB-GENO-100908-4,ZFIN:ZDB-MRPHLNO-060113-4 | Zfish | PMID:19307601 |
| Biological Process | GO:1904753 | negative regulation of vascular associated smooth muscle cell migration | Gna12 | IMP | | Rat | PMID:29453251|RGD:42721986 |
| Biological Process | GO:1904706 | negative regulation of vascular associated smooth muscle cell proliferation | Gna12 | IMP | | Rat | PMID:29453251|RGD:42721986 |
| Biological Process | GO:0014003 | oligodendrocyte development | gna12a | IGI | ZFIN:ZDB-GENO-160202-2,ZFIN:ZDB-MRPHLNO-060113-1,ZFIN:ZDB-MRPHLNO-060113-3,ZFIN:ZDB-MRPHLNO-060113-4 | Zfish | PMID:25607772 |
| Biological Process | GO:0032516 | positive regulation of phosphoprotein phosphatase activity | GNA12 | IDA | | Human | PMID:15525651 |
| Biological Process | GO:0032956 | regulation of actin cytoskeleton organization | gna12a | IGI | ZFIN:ZDB-MRPHLNO-060113-1,ZFIN:ZDB-MRPHLNO-060113-3,ZFIN:ZDB-MRPHLNO-060113-4 | Zfish | PMID:19307601 |
| Biological Process | GO:0008217 | regulation of blood pressure | Gna12 | IGI | MGI:MGI:95768 | Mouse | PMID:26531127 |
| Biological Process | GO:0008360 | regulation of cell shape | Gna12 | IDA | | Mouse | PMID:10037795 |
| Biological Process | GO:0010762 | regulation of fibroblast migration | Gna12 | IMP | | Mouse | MGI:MGI:5448916|PMID:22609986 |
| Biological Process | GO:0032434 | regulation of proteasomal ubiquitin-dependent protein catabolic process | GNA12 | IMP | | Human | PMID:22609986 |
| Biological Process | GO:0032434 | regulation of proteasomal ubiquitin-dependent protein catabolic process | Gna12 | IMP | | Mouse | MGI:MGI:5448916|PMID:22609986 |
| Biological Process | GO:0032006 | regulation of TOR signaling | GNA12 | IMP | | Human | PMID:22609986 |
| Biological Process | GO:0032006 | regulation of TOR signaling | Gna12 | IMP | | Mouse | MGI:MGI:5448916|PMID:22609986 |
| Biological Process | GO:0009410 | response to xenobiotic stimulus | Gna12 | IEP | | Rat | PMID:12623966|RGD:730287 |
| Biological Process | GO:0007266 | Rho protein signal transduction | Gna12 | IDA | | Mouse | PMID:10037795 |
| Biological Process | GO:0007266 | Rho protein signal transduction | gna12a | IMP | | Zfish | PMID:15928205 |
| Biological Process | GO:0043149 | stress fiber assembly | gna12a | IMP | | Zfish | PMID:15928205 |
| Biological Process | GO:0009826 | unidimensional cell growth | gna12a | IGI | ZFIN:ZDB-MRPHLNO-060113-1,ZFIN:ZDB-MRPHLNO-060113-3,ZFIN:ZDB-MRPHLNO-060113-4 | Zfish | PMID:15928205 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
 Analysis Tools
Analysis Tools