|
Source |
Alliance of Genome Resources |
| Category | ID | Classification term | Gene | Evidence | Inferred from | Organism | Reference |
|---|---|---|---|---|---|---|---|
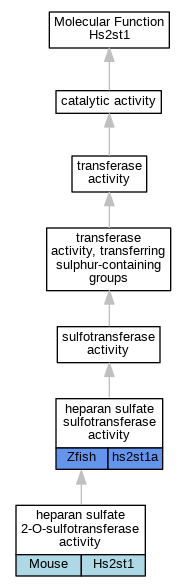
| Molecular Function | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity | Hs2st1 | IMP | MGI:MGI:4439258 | Mouse | PMID:19889634 |
| Molecular Function | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity | Hs2st1 | IDA | Mouse | PMID:15226404 | |
| Molecular Function | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity | Hs2st1 | IMP | MGI:MGI:1934916 | Mouse | PMID:11457822 |
| Molecular Function | GO:0034483 | heparan sulfate sulfotransferase activity | hs2st1a | IMP | ZFIN:ZDB-MRPHLNO-120420-2 | Zfish | PMID:22357927 |
| Molecular Function | GO:0034483 | heparan sulfate sulfotransferase activity | hs2st1a | IMP | ZFIN:ZDB-MRPHLNO-120420-1 | Zfish | PMID:22357927 |
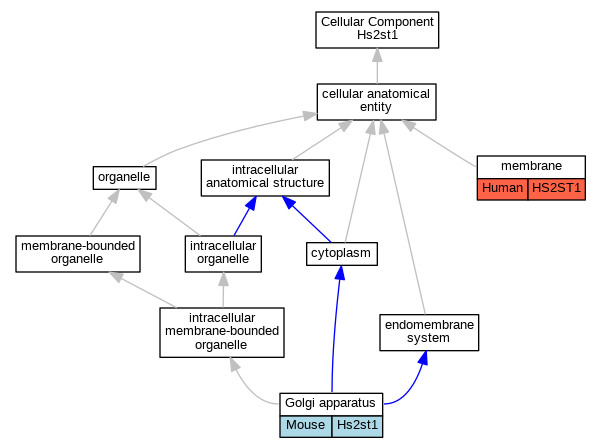
| Cellular Component | GO:0005794 | Golgi apparatus | Hs2st1 | IDA | Mouse | PMID:24326668 | |
| Cellular Component | GO:0005794 | Golgi apparatus | Hs2st1 | IDA | Mouse | PMID:11687650 | |
| Cellular Component | GO:0005794 | Golgi apparatus | Hs2st1 | IDA | Mouse | PMID:15226404 | |
| Cellular Component | GO:0016020 | membrane | HS2ST1 | HDA | Human | PMID:19946888 | |
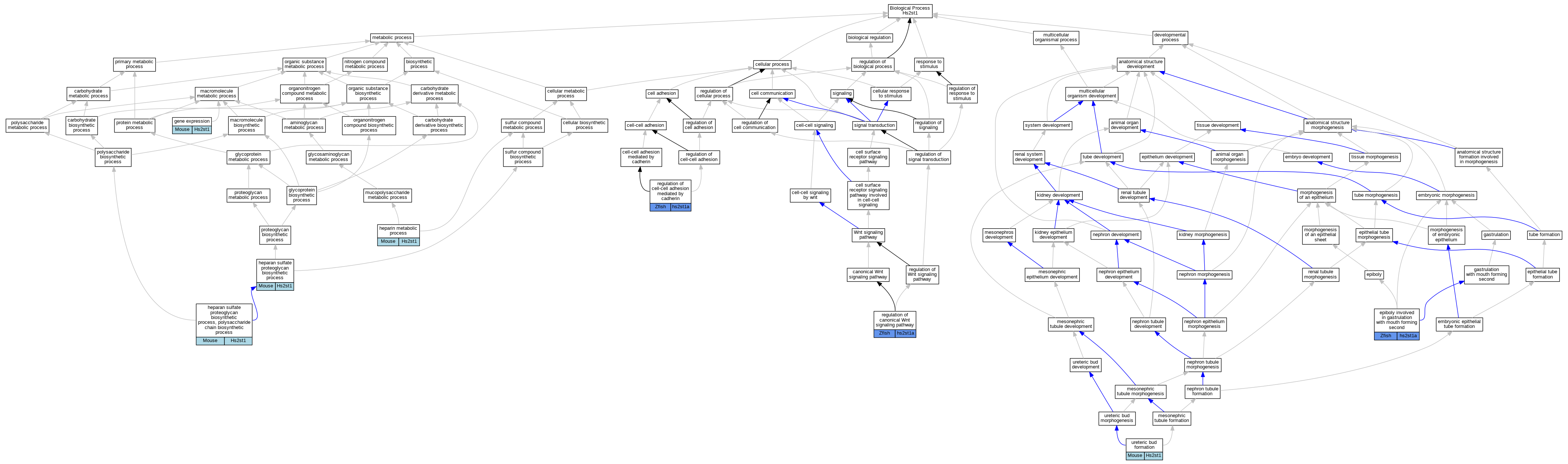
| Biological Process | GO:0055113 | epiboly involved in gastrulation with mouth forming second | hs2st1a | IMP | ZFIN:ZDB-MRPHLNO-120420-2 | Zfish | PMID:22357927 |
| Biological Process | GO:0055113 | epiboly involved in gastrulation with mouth forming second | hs2st1a | IMP | ZFIN:ZDB-MRPHLNO-120420-1 | Zfish | PMID:22357927 |
| Biological Process | GO:0010467 | gene expression | Hs2st1 | IMP | Mouse | PMID:32201365 | |
| Biological Process | GO:0015012 | heparan sulfate proteoglycan biosynthetic process | Hs2st1 | IMP | MGI:MGI:4439258 | Mouse | PMID:19889634 |
| Biological Process | GO:0015012 | heparan sulfate proteoglycan biosynthetic process | Hs2st1 | IMP | Mouse | PMID:30377379 | |
| Biological Process | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process | Hs2st1 | IMP | MGI:MGI:1934916 | Mouse | PMID:11457822 |
| Biological Process | GO:0030202 | heparin metabolic process | Hs2st1 | IMP | MGI:MGI:1934916 | Mouse | PMID:11457822 |
| Biological Process | GO:0060828 | regulation of canonical Wnt signaling pathway | hs2st1a | IMP | ZFIN:ZDB-MRPHLNO-120420-2 | Zfish | PMID:22357927 |
| Biological Process | GO:0060828 | regulation of canonical Wnt signaling pathway | hs2st1a | IMP | ZFIN:ZDB-MRPHLNO-120420-1 | Zfish | PMID:22357927 |
| Biological Process | GO:2000047 | regulation of cell-cell adhesion mediated by cadherin | hs2st1a | IMP | ZFIN:ZDB-MRPHLNO-120420-1 | Zfish | PMID:22357927 |
| Biological Process | GO:2000047 | regulation of cell-cell adhesion mediated by cadherin | hs2st1a | IMP | ZFIN:ZDB-MRPHLNO-120420-2 | Zfish | PMID:22357927 |
| Biological Process | GO:0060676 | ureteric bud formation | Hs2st1 | IMP | MGI:MGI:1934916 | Mouse | PMID:9637690 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 07/05/2024 MGI 6.24 |

|
|
|
||