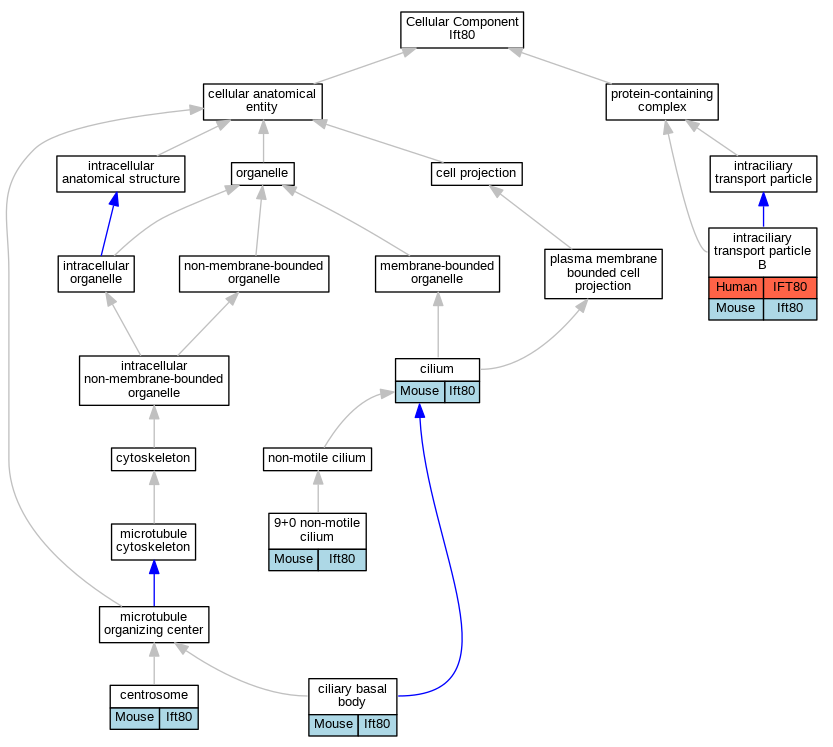
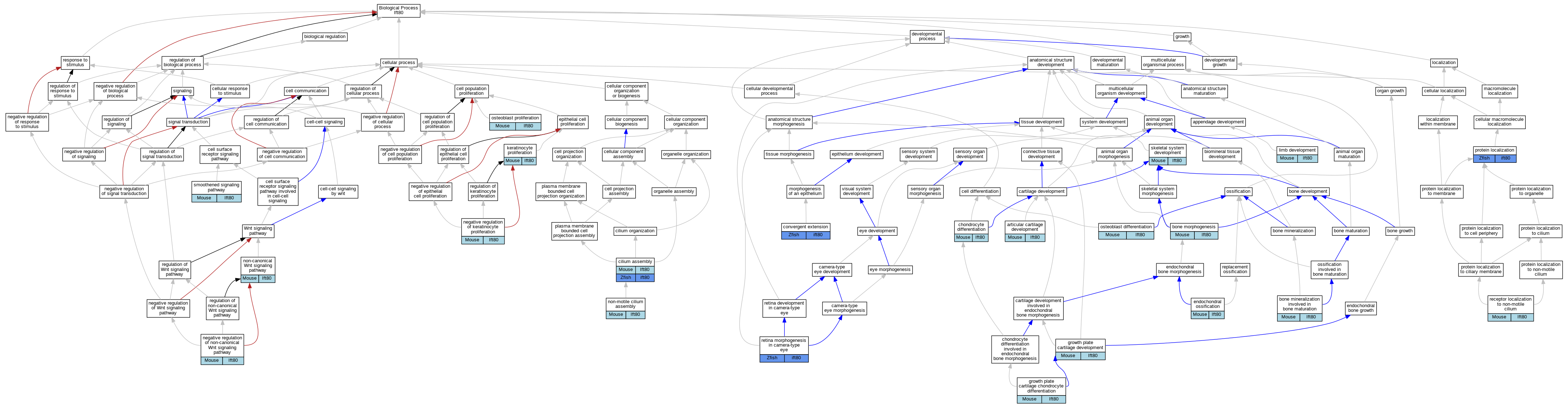
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Cellular Component | GO:0097731 | 9+0 non-motile cilium | Ift80 | IDA | | Mouse | PMID:26996322 |
| Cellular Component | GO:0005813 | centrosome | Ift80 | IDA | | Mouse | PMID:19253336 |
| Cellular Component | GO:0036064 | ciliary basal body | Ift80 | IDA | | Mouse | PMID:26996322 |
| Cellular Component | GO:0005929 | cilium | Ift80 | IDA | | Mouse | PMID:19253336 |
| Cellular Component | GO:0005929 | cilium | Ift80 | IDA | | Mouse | PMID:23333501 |
| Cellular Component | GO:0030992 | intraciliary transport particle B | IFT80 | IPI | | Human | PMID:26980730 |
| Cellular Component | GO:0030992 | intraciliary transport particle B | Ift80 | IDA | | Mouse | MGI:MGI:5607391|PMID:24596149 |
| Cellular Component | GO:0030992 | intraciliary transport particle B | Ift80 | IDA | | Mouse | MGI:MGI:5613829|PMID:25443296 |
| Cellular Component | GO:0030992 | intraciliary transport particle B | Ift80 | IDA | | Mouse | PMID:19253336 |
| Cellular Component | GO:0030992 | intraciliary transport particle B | Ift80 | IDA | | Mouse | PMID:23810713 |
| Biological Process | GO:0061975 | articular cartilage development | Ift80 | IMP | MGI:5818514,MGI:3665440 | Mouse | PMID:26098911 |
| Biological Process | GO:0035630 | bone mineralization involved in bone maturation | Ift80 | IMP | MGI:5818514,MGI:3689350 | Mouse | PMID:26996322 |
| Biological Process | GO:0060349 | bone morphogenesis | Ift80 | IMP | MGI:MGI:3856827 | Mouse | PMID:21227999 |
| Biological Process | GO:0002062 | chondrocyte differentiation | Ift80 | IGI | MGI:MGI:95728 | Mouse | PMID:23333501 |
| Biological Process | GO:0002062 | chondrocyte differentiation | Ift80 | IMP | | Mouse | PMID:23333501 |
| Biological Process | GO:0060271 | cilium assembly | Ift80 | IMP | | Mouse | PMID:23333501 |
| Biological Process | GO:0060271 | cilium assembly | ift80 | IMP | ZFIN:ZDB-MRPHLNO-070605-8 | Zfish | PMID:20207966 |
| Biological Process | GO:0060026 | convergent extension | ift80 | IGI | ZFIN:ZDB-MRPHLNO-060301-2,ZFIN:ZDB-MRPHLNO-070605-8 | Zfish | PMID:20207966 |
| Biological Process | GO:0060026 | convergent extension | ift80 | IGI | ZFIN:ZDB-MRPHLNO-060208-1,ZFIN:ZDB-MRPHLNO-070605-8 | Zfish | PMID:20207966 |
| Biological Process | GO:0001958 | endochondral ossification | Ift80 | IMP | MGI:5818514,MGI:3665440 | Mouse | PMID:26098911 |
| Biological Process | GO:0003418 | growth plate cartilage chondrocyte differentiation | Ift80 | IMP | MGI:5818514,MGI:3665440 | Mouse | PMID:26098911 |
| Biological Process | GO:0003417 | growth plate cartilage development | Ift80 | IMP | MGI:5818514,MGI:3665440 | Mouse | PMID:26098911 |
| Biological Process | GO:0043616 | keratinocyte proliferation | Ift80 | IMP | | Mouse | PMID:21703454 |
| Biological Process | GO:0060173 | limb development | Ift80 | IMP | MGI:5818514,MGI:3665440 | Mouse | PMID:26098911 |
| Biological Process | GO:0010839 | negative regulation of keratinocyte proliferation | Ift80 | IMP | | Mouse | PMID:21703454 |
| Biological Process | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway | Ift80 | IGI | MGI:MGI:98956 | Mouse | PMID:23333501 |
| Biological Process | GO:0035567 | non-canonical Wnt signaling pathway | Ift80 | IGI | MGI:MGI:98956 | Mouse | PMID:23333501 |
| Biological Process | GO:1905515 | non-motile cilium assembly | Ift80 | IMP | MGI:5818514,MGI:3665440 | Mouse | PMID:26098911 |
| Biological Process | GO:1905515 | non-motile cilium assembly | Ift80 | IMP | MGI:5818514,MGI:3689350 | Mouse | PMID:26996322 |
| Biological Process | GO:1905515 | non-motile cilium assembly | Ift80 | IMP | | Mouse | PMID:24927541 |
| Biological Process | GO:1905515 | non-motile cilium assembly | Ift80 | IMP | | Mouse | PMID:21703454 |
| Biological Process | GO:1905515 | non-motile cilium assembly | Ift80 | IMP | | Mouse | PMID:22771375 |
| Biological Process | GO:0001649 | osteoblast differentiation | Ift80 | IMP | MGI:5818514,MGI:3689350 | Mouse | PMID:26996322 |
| Biological Process | GO:0001649 | osteoblast differentiation | Ift80 | IGI | MGI:MGI:95728 | Mouse | PMID:22771375 |
| Biological Process | GO:0001649 | osteoblast differentiation | Ift80 | IMP | | Mouse | PMID:22771375 |
| Biological Process | GO:0033687 | osteoblast proliferation | Ift80 | IMP | MGI:5818514,MGI:3689350 | Mouse | PMID:26996322 |
| Biological Process | GO:0008104 | protein localization | ift80 | IMP | ZFIN:ZDB-MRPHLNO-070605-8 | Zfish | PMID:20207966 |
| Biological Process | GO:0097500 | receptor localization to non-motile cilium | Ift80 | IMP | MGI:5818514,MGI:3689350 | Mouse | PMID:26996322 |
| Biological Process | GO:0060042 | retina morphogenesis in camera-type eye | ift80 | IMP | ZFIN:ZDB-MRPHLNO-070605-8 | Zfish | PMID:20207966 |
| Biological Process | GO:0001501 | skeletal system development | Ift80 | IMP | MGI:5818514,MGI:3689350 | Mouse | PMID:26996322 |
| Biological Process | GO:0007224 | smoothened signaling pathway | Ift80 | IMP | MGI:5818514,MGI:3665440 | Mouse | PMID:26098911 |
| Biological Process | GO:0007224 | smoothened signaling pathway | Ift80 | IMP | MGI:5818514,MGI:3689350 | Mouse | PMID:26996322 |
| Biological Process | GO:0007224 | smoothened signaling pathway | Ift80 | IMP | MGI:MGI:3856827 | Mouse | PMID:21227999 |
| Biological Process | GO:0007224 | smoothened signaling pathway | Ift80 | IMP | | Mouse | PMID:23333501 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
 Analysis Tools
Analysis Tools