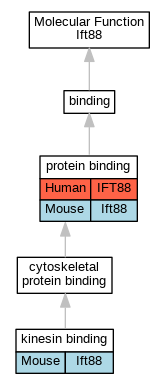
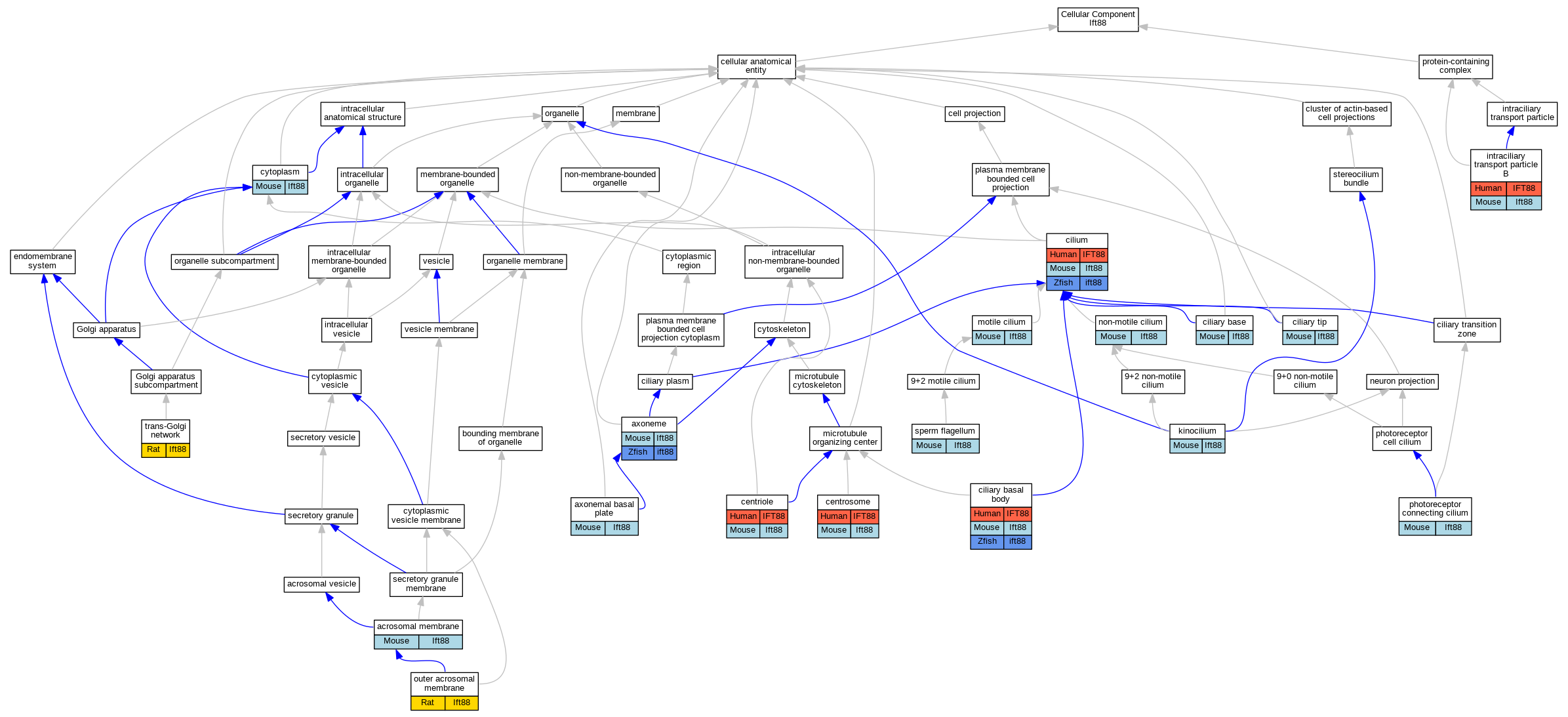
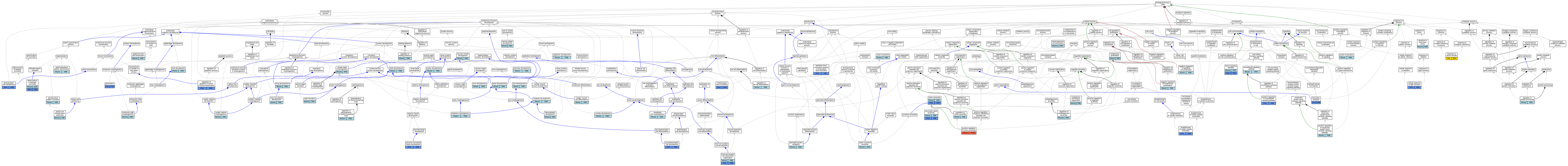
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0019894 | kinesin binding | Ift88 | IPI | PR:Q99PW8 | Mouse | PMID:19384852 |
| Molecular Function | GO:0005515 | protein binding | IFT88 | IPI | UniProtKB:P49454 | Human | PMID:25564561 |
| Molecular Function | GO:0005515 | protein binding | IFT88 | IPI | UniProtKB:Q96C92 | Human | PMID:27767179 |
| Molecular Function | GO:0005515 | protein binding | IFT88 | IPI | UniProtKB:Q8IYG6 | Human | PMID:30388400 |
| Molecular Function | GO:0005515 | protein binding | IFT88 | IPI | UniProtKB:A0A0C4DGQ7 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | IFT88 | IPI | UniProtKB:P27987 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | IFT88 | IPI | UniProtKB:Q969G3 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | IFT88 | IPI | UniProtKB:Q96G04 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | IFT88 | IPI | UniProtKB:Q96HB5 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | IFT88 | IPI | UniProtKB:Q9NZV6 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | IFT88 | IPI | UniProtKB:Q9Y6W3 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | Ift88 | IPI | UniProtKB:Q6VH22 | Mouse | MGI:MGI:1926671|PMID:11062270 |
| Molecular Function | GO:0005515 | protein binding | Ift88 | IPI | PR:Q52KB6 | Mouse | PMID:24469809 |
| Molecular Function | GO:0005515 | protein binding | Ift88 | IPI | PR:Q9DB07 | Mouse | PMID:17312020 |
| Cellular Component | GO:0002080 | acrosomal membrane | Ift88 | IDA | | Mouse | PMID:21337470 |
| Cellular Component | GO:0097541 | axonemal basal plate | Ift88 | IDA | | Mouse | PMID:24302887 |
| Cellular Component | GO:0005930 | axoneme | Ift88 | IDA | | Mouse | PMID:11251073 |
| Cellular Component | GO:0005930 | axoneme | Ift88 | IDA | | Mouse | PMID:24302887 |
| Cellular Component | GO:0005930 | axoneme | Ift88 | IDA | | Mouse | PMID:18066062 |
| Cellular Component | GO:0005930 | axoneme | Ift88 | IDA | | Mouse | PMID:20368623 |
| Cellular Component | GO:0005930 | axoneme | Ift88 | IDA | | Mouse | PMID:11854326 |
| Cellular Component | GO:0005930 | axoneme | ift88 | IDA | | Zfish | PMID:18304522 |
| Cellular Component | GO:0005814 | centriole | IFT88 | IDA | | Human | PMID:27623382 |
| Cellular Component | GO:0005814 | centriole | Ift88 | IDA | | Mouse | MGI:MGI:4457459|PMID:20230748 |
| Cellular Component | GO:0005814 | centriole | Ift88 | IDA | | Mouse | MGI:MGI:5559658|PMID:23386061 |
| Cellular Component | GO:0005814 | centriole | Ift88 | IDA | | Mouse | PMID:29038301 |
| Cellular Component | GO:0005814 | centriole | Ift88 | IDA | | Mouse | PMID:24469809 |
| Cellular Component | GO:0005814 | centriole | Ift88 | IDA | | Mouse | PMID:29487109 |
| Cellular Component | GO:0005813 | centrosome | IFT88 | IDA | | Human | PMID:27767179 |
| Cellular Component | GO:0005813 | centrosome | Ift88 | IDA | | Mouse | PMID:19253336 |
| Cellular Component | GO:0005813 | centrosome | Ift88 | IDA | | Mouse | PMID:25564561 |
| Cellular Component | GO:0036064 | ciliary basal body | IFT88 | IDA | | Human | PMID:17604723 |
| Cellular Component | GO:0036064 | ciliary basal body | IFT88 | IDA | | Human | PMID:27767179 |
| Cellular Component | GO:0036064 | ciliary basal body | IFT88 | IDA | | Human | PMID:28428259 |
| Cellular Component | GO:0036064 | ciliary basal body | IFT88 | IDA | | Human | PMID:28625565 |
| Cellular Component | GO:0036064 | ciliary basal body | Ift88 | IDA | | Mouse | MGI:MGI:4457459|PMID:20230748 |
| Cellular Component | GO:0036064 | ciliary basal body | Ift88 | IDA | | Mouse | PMID:11854326 |
| Cellular Component | GO:0036064 | ciliary basal body | Ift88 | IDA | | Mouse | PMID:11251073 |
| Cellular Component | GO:0036064 | ciliary basal body | Ift88 | IDA | | Mouse | PMID:23955340 |
| Cellular Component | GO:0036064 | ciliary basal body | ift88 | IDA | | Zfish | PMID:18492793 |
| Cellular Component | GO:0036064 | ciliary basal body | ift88 | IDA | | Zfish | PMID:18304522 |
| Cellular Component | GO:0097546 | ciliary base | Ift88 | IDA | | Mouse | PMID:16775004 |
| Cellular Component | GO:0097546 | ciliary base | Ift88 | IDA | | Mouse | PMID:19654211 |
| Cellular Component | GO:0097546 | ciliary base | Ift88 | IDA | | Mouse | PMID:21653639 |
| Cellular Component | GO:0097542 | ciliary tip | Ift88 | IDA | | Mouse | PMID:19654211 |
| Cellular Component | GO:0097542 | ciliary tip | Ift88 | IDA | | Mouse | PMID:21653639 |
| Cellular Component | GO:0097542 | ciliary tip | Ift88 | IDA | | Mouse | PMID:16775004 |
| Cellular Component | GO:0097542 | ciliary tip | Ift88 | IDA | | Mouse | PMID:23955340 |
| Cellular Component | GO:0005929 | cilium | IFT88 | IDA | | Human | PMID:24421332 |
| Cellular Component | GO:0005929 | cilium | IFT88 | IDA | | Human | PMID:27727273 |
| Cellular Component | GO:0005929 | cilium | IFT88 | IDA | | Human | PMID:28428259 |
| Cellular Component | GO:0005929 | cilium | Ift88 | IDA | | Mouse | MGI:MGI:4888042|PMID:21209331 |
| Cellular Component | GO:0005929 | cilium | Ift88 | IDA | | Mouse | MGI:MGI:4457459|PMID:20230748 |
| Cellular Component | GO:0005929 | cilium | Ift88 | IDA | | Mouse | PMID:24469809 |
| Cellular Component | GO:0005929 | cilium | Ift88 | IDA | | Mouse | PMID:29487109 |
| Cellular Component | GO:0005929 | cilium | Ift88 | IDA | | Mouse | PMID:19253336 |
| Cellular Component | GO:0005929 | cilium | Ift88 | IDA | | Mouse | PMID:20159594 |
| Cellular Component | GO:0005929 | cilium | Ift88 | IDA | | Mouse | PMID:27727273 |
| Cellular Component | GO:0005929 | cilium | Ift88 | IDA | | Mouse | PMID:24927541 |
| Cellular Component | GO:0005929 | cilium | Ift88 | IDA | | Mouse | PMID:18285569 |
| Cellular Component | GO:0005929 | cilium | Ift88 | IDA | | Mouse | PMID:22689656 |
| Cellular Component | GO:0005929 | cilium | Ift88 | IDA | | Mouse | PMID:25294941 |
| Cellular Component | GO:0005929 | cilium | ift88 | IDA | | Zfish | PMID:16216239 |
| Cellular Component | GO:0005737 | cytoplasm | Ift88 | IDA | | Mouse | MGI:MGI:6201947|PMID:27682589 |
| Cellular Component | GO:0030992 | intraciliary transport particle B | IFT88 | IPI | | Human | PMID:26980730 |
| Cellular Component | GO:0030992 | intraciliary transport particle B | Ift88 | IDA | | Mouse | MGI:MGI:5613829|PMID:25443296 |
| Cellular Component | GO:0030992 | intraciliary transport particle B | Ift88 | IDA | | Mouse | MGI:MGI:5607391|PMID:24596149 |
| Cellular Component | GO:0030992 | intraciliary transport particle B | Ift88 | IDA | | Mouse | PMID:12821668 |
| Cellular Component | GO:0030992 | intraciliary transport particle B | Ift88 | IDA | | Mouse | PMID:19253336 |
| Cellular Component | GO:0030992 | intraciliary transport particle B | Ift88 | IDA | | Mouse | PMID:23810713 |
| Cellular Component | GO:0060091 | kinocilium | Ift88 | IDA | | Mouse | PMID:18066062 |
| Cellular Component | GO:0031514 | motile cilium | Ift88 | IDA | | Mouse | PMID:18590716 |
| Cellular Component | GO:0097730 | non-motile cilium | Ift88 | IDA | | Mouse | PMID:16254602 |
| Cellular Component | GO:0002081 | outer acrosomal membrane | Ift88 | IDA | | Rat | PMID:21337470|RGD:8655529 |
| Cellular Component | GO:0032391 | photoreceptor connecting cilium | Ift88 | IDA | | Mouse | MGI:MGI:3839506|PMID:19208653 |
| Cellular Component | GO:0032391 | photoreceptor connecting cilium | Ift88 | IDA | | Mouse | PMID:20368623 |
| Cellular Component | GO:0036126 | sperm flagellum | Ift88 | IDA | | Mouse | MGI:MGI:6201947|PMID:27682589 |
| Cellular Component | GO:0005802 | trans-Golgi network | Ift88 | IDA | | Rat | PMID:21337470|RGD:8655529 |
| Biological Process | GO:0009887 | animal organ morphogenesis | Ift88 | IMP | MGI:MGI:2157527 | Mouse | PMID:15226261 |
| Biological Process | GO:0009952 | anterior/posterior pattern specification | Ift88 | IMP | MGI:MGI:3027342 | Mouse | PMID:15930098 |
| Biological Process | GO:0009952 | anterior/posterior pattern specification | Ift88 | IMP | MGI:MGI:2157527 | Mouse | PMID:12701101 |
| Biological Process | GO:0007420 | brain development | Ift88 | IMP | MGI:MGI:3823090 | Mouse | PMID:19036983 |
| Biological Process | GO:0007420 | brain development | Ift88 | IGI | MGI:MGI:1918742 | Mouse | PMID:22228099 |
| Biological Process | GO:0055007 | cardiac muscle cell differentiation | Ift88 | IMP | MGI:MGI:1859153 | Mouse | PMID:19654211 |
| Biological Process | GO:0055007 | cardiac muscle cell differentiation | Ift88 | IMP | | Mouse | PMID:19654211 |
| Biological Process | GO:0060411 | cardiac septum morphogenesis | Ift88 | IMP | MGI:MGI:3823090 | Mouse | PMID:19036983 |
| Biological Process | GO:0071482 | cellular response to light stimulus | ift88 | IMP | ZFIN:ZDB-GENO-080730-2 | Zfish | PMID:22485131 |
| Biological Process | GO:0090660 | cerebrospinal fluid circulation | ift88 | IMP | ZFIN:ZDB-GENO-080730-2 | Zfish | PMID:30612902 |
| Biological Process | GO:0060271 | cilium assembly | Ift88 | IMP | | Mouse | MGI:MGI:6383338|PMID:31761534 |
| Biological Process | GO:0060271 | cilium assembly | Ift88 | IMP | | Mouse | MGI:MGI:1926671|PMID:11062270 |
| Biological Process | GO:0060271 | cilium assembly | Ift88 | IMP | | Mouse | PMID:19654211 |
| Biological Process | GO:0060271 | cilium assembly | Ift88 | IMP | | Mouse | PMID:21565611 |
| Biological Process | GO:0060271 | cilium assembly | Ift88 | IMP | | Mouse | PMID:21289087 |
| Biological Process | GO:0060271 | cilium assembly | ift88 | IMP | ZFIN:ZDB-MRPHLNO-050506-1 | Zfish | PMID:31015398 |
| Biological Process | GO:0060271 | cilium assembly | ift88 | IMP | ZFIN:ZDB-GENO-080730-2 | Zfish | PMID:30612902 |
| Biological Process | GO:0060271 | cilium assembly | ift88 | IMP | ZFIN:ZDB-GENO-080730-2 | Zfish | PMID:18364699 |
| Biological Process | GO:0060271 | cilium assembly | ift88 | IMP | ZFIN:ZDB-MRPHLNO-050506-1 | Zfish | PMID:15790966 |
| Biological Process | GO:0060271 | cilium assembly | ift88 | IMP | ZFIN:ZDB-GENO-160908-10 | Zfish | PMID:27207957 |
| Biological Process | GO:0060271 | cilium assembly | ift88 | IMP | ZFIN:ZDB-MRPHLNO-050506-1 | Zfish | PMID:24743595 |
| Biological Process | GO:0060271 | cilium assembly | ift88 | IMP | ZFIN:ZDB-GENO-110203-15 | Zfish | PMID:19700616 |
| Biological Process | GO:0060271 | cilium assembly | ift88 | IMP | ZFIN:ZDB-GENO-080730-2 | Zfish | PMID:19517571 |
| Biological Process | GO:0060271 | cilium assembly | ift88 | IMP | ZFIN:ZDB-MRPHLNO-060111-1 | Zfish | PMID:16216239 |
| Biological Process | GO:0060271 | cilium assembly | ift88 | IMP | ZFIN:ZDB-GENO-980410-147 | Zfish | PMID:15790966 |
| Biological Process | GO:0060271 | cilium assembly | ift88 | IMP | ZFIN:ZDB-MRPHLNO-050506-2 | Zfish | PMID:25838181 |
| Biological Process | GO:0060271 | cilium assembly | ift88 | IMP | ZFIN:ZDB-MRPHLNO-050513-1 | Zfish | PMID:19517571 |
| Biological Process | GO:0060271 | cilium assembly | ift88 | IMP | ZFIN:ZDB-GENO-080730-2 | Zfish | PMID:18492793 |
| Biological Process | GO:0003341 | cilium movement | Ift88 | IMP | MGI:3710185 | Mouse | PMID:29891944 |
| Biological Process | GO:0090102 | cochlea development | Ift88 | IGI | MGI:MGI:2135272 | Mouse | PMID:18066062 |
| Biological Process | GO:0031122 | cytoplasmic microtubule organization | Ift88 | IMP | | Mouse | PMID:21703454 |
| Biological Process | GO:0007368 | determination of left/right symmetry | Ift88 | IMP | MGI:MGI:3027342 | Mouse | PMID:15755804 |
| Biological Process | GO:0007368 | determination of left/right symmetry | Ift88 | IMP | MGI:MGI:1859153 | Mouse | PMID:10804177 |
| Biological Process | GO:0007368 | determination of left/right symmetry | ift88 | IGI | ZFIN:ZDB-GENO-080301-1,ZFIN:ZDB-MRPHLNO-050830-1 | Zfish | PMID:18056639 |
| Biological Process | GO:0007368 | determination of left/right symmetry | ift88 | IGI | ZFIN:ZDB-MRPHLNO-050506-1,ZFIN:ZDB-MRPHLNO-050830-1 | Zfish | PMID:18056639 |
| Biological Process | GO:0007368 | determination of left/right symmetry | ift88 | IMP | ZFIN:ZDB-MRPHLNO-060111-1 | Zfish | PMID:16216239 |
| Biological Process | GO:0009953 | dorsal/ventral pattern formation | Ift88 | IMP | MGI:MGI:3027342 | Mouse | PMID:15755804 |
| Biological Process | GO:0009953 | dorsal/ventral pattern formation | Ift88 | IMP | MGI:MGI:3027342 | Mouse | PMID:15930098 |
| Biological Process | GO:0042733 | embryonic digit morphogenesis | Ift88 | IMP | MGI:MGI:3027342 | Mouse | PMID:15930098 |
| Biological Process | GO:0042733 | embryonic digit morphogenesis | Ift88 | IGI | MGI:MGI:98297 | Mouse | PMID:15930098 |
| Biological Process | GO:0042733 | embryonic digit morphogenesis | Ift88 | IMP | MGI:MGI:3027342 | Mouse | PMID:15755804 |
| Biological Process | GO:0042733 | embryonic digit morphogenesis | Ift88 | IMP | MGI:MGI:2157527 | Mouse | PMID:16254602 |
| Biological Process | GO:0042733 | embryonic digit morphogenesis | Ift88 | IMP | MGI:MGI:2157527 | Mouse | PMID:12701101 |
| Biological Process | GO:0042733 | embryonic digit morphogenesis | Ift88 | IMP | MGI:MGI:3823090 | Mouse | PMID:19036983 |
| Biological Process | GO:0001886 | endothelial cell morphogenesis | Ift88 | IMP | MGI:MGI:2157527 | Mouse | PMID:21285373 |
| Biological Process | GO:0036334 | epidermal stem cell homeostasis | Ift88 | IMP | MGI:MGI:2445832,MGI:MGI:3710185 | Mouse | PMID:21429982 |
| Biological Process | GO:0008544 | epidermis development | Ift88 | IMP | MGI:MGI:1926500,MGI:MGI:3710185 | Mouse | PMID:21703454 |
| Biological Process | GO:0003351 | epithelial cilium movement involved in extracellular fluid movement | ift88 | IMP | ZFIN:ZDB-GENO-980410-147 | Zfish | PMID:15790966 |
| Biological Process | GO:0003351 | epithelial cilium movement involved in extracellular fluid movement | ift88 | IMP | ZFIN:ZDB-MRPHLNO-050506-1 | Zfish | PMID:15790966 |
| Biological Process | GO:0000132 | establishment of mitotic spindle orientation | ift88 | IMP | ZFIN:ZDB-GENO-120510-4 | Zfish | PMID:24095732 |
| Biological Process | GO:0000132 | establishment of mitotic spindle orientation | ift88 | IMP | ZFIN:ZDB-MRPHLNO-050506-1 | Zfish | PMID:21441926 |
| Biological Process | GO:0001654 | eye development | Ift88 | IGI | MGI:MGI:1918742 | Mouse | PMID:22228099 |
| Biological Process | GO:0042462 | eye photoreceptor cell development | ift88 | IMP | ZFIN:ZDB-GENO-080730-2 | Zfish | PMID:18492793 |
| Biological Process | GO:0042462 | eye photoreceptor cell development | ift88 | IMP | ZFIN:ZDB-GENO-980410-147 | Zfish | PMID:15182712 |
| Biological Process | GO:0042462 | eye photoreceptor cell development | ift88 | IMP | ZFIN:ZDB-MRPHLNO-050513-1 | Zfish | PMID:18492793 |
| Biological Process | GO:0030900 | forebrain development | Ift88 | IMP | MGI:MGI:1932522,MGI:MGI:3710185 | Mouse | PMID:28291836 |
| Biological Process | GO:0048853 | forebrain morphogenesis | Ift88 | IMP | MGI:MGI:3823090 | Mouse | PMID:19036983 |
| Biological Process | GO:0007369 | gastrulation | ift88 | IMP | ZFIN:ZDB-MRPHLNO-050513-2 | Zfish | PMID:22941275 |
| Biological Process | GO:0007507 | heart development | Ift88 | IGI | MGI:MGI:1918742 | Mouse | PMID:22228099 |
| Biological Process | GO:0060914 | heart formation | Ift88 | IMP | MGI:MGI:1859153 | Mouse | PMID:19654211 |
| Biological Process | GO:0060122 | inner ear receptor cell stereocilium organization | Ift88 | IGI | MGI:MGI:2135272 | Mouse | PMID:18066062 |
| Biological Process | GO:0060122 | inner ear receptor cell stereocilium organization | Ift88 | IMP | MGI:MGI:1932522,MGI:MGI:3710185 | Mouse | PMID:18066062 |
| Biological Process | GO:0060122 | inner ear receptor cell stereocilium organization | ift88 | IMP | | Zfish | PMID:24626987 |
| Biological Process | GO:0042073 | intraciliary transport | ift88 | IMP | ZFIN:ZDB-GENO-080730-2 | Zfish | PMID:18492793 |
| Biological Process | GO:0042073 | intraciliary transport | ift88 | IMP | ZFIN:ZDB-MRPHLNO-050513-1 | Zfish | PMID:18492793 |
| Biological Process | GO:0043616 | keratinocyte proliferation | Ift88 | IMP | | Mouse | PMID:21703454 |
| Biological Process | GO:0043616 | keratinocyte proliferation | Ift88 | IMP | MGI:MGI:1926500,MGI:MGI:3710185 | Mouse | PMID:21703454 |
| Biological Process | GO:0001822 | kidney development | Ift88 | IMP | MGI:MGI:2157527 | Mouse | PMID:9176412 |
| Biological Process | GO:0001822 | kidney development | Ift88 | IMP | MGI:MGI:2157527 | Mouse | PMID:11773599 |
| Biological Process | GO:0060173 | limb development | Ift88 | IGI | MGI:MGI:1918742 | Mouse | PMID:22228099 |
| Biological Process | GO:0001889 | liver development | Ift88 | IMP | MGI:MGI:2157527 | Mouse | PMID:9176412 |
| Biological Process | GO:0001889 | liver development | Ift88 | IMP | MGI:MGI:2157527 | Mouse | PMID:11773599 |
| Biological Process | GO:0030324 | lung development | Ift88 | IMP | MGI:MGI:3823090 | Mouse | PMID:19036983 |
| Biological Process | GO:0060426 | lung vasculature development | Ift88 | IMP | MGI:MGI:3823090 | Mouse | PMID:19036983 |
| Biological Process | GO:0050680 | negative regulation of epithelial cell proliferation | Ift88 | IMP | MGI:MGI:1926500,MGI:MGI:3710185 | Mouse | PMID:21703454 |
| Biological Process | GO:0010839 | negative regulation of keratinocyte proliferation | Ift88 | IMP | | Mouse | PMID:21703454 |
| Biological Process | GO:0010839 | negative regulation of keratinocyte proliferation | Ift88 | IMP | MGI:MGI:1926500,MGI:MGI:3710185 | Mouse | PMID:21703454 |
| Biological Process | GO:0007399 | nervous system development | Ift88 | IMP | MGI:MGI:3823090 | Mouse | PMID:19036983 |
| Biological Process | GO:0061351 | neural precursor cell proliferation | Ift88 | IMP | MGI:MGI:2157527 | Mouse | PMID:18297065 |
| Biological Process | GO:1905515 | non-motile cilium assembly | Ift88 | IMP | | Mouse | PMID:21703454 |
| Biological Process | GO:1905515 | non-motile cilium assembly | Ift88 | IMP | MGI:MGI:2157527 | Mouse | PMID:21285373 |
| Biological Process | GO:1905515 | non-motile cilium assembly | Ift88 | IMP | | Mouse | PMID:24927541 |
| Biological Process | GO:1905515 | non-motile cilium assembly | Ift88 | IMP | MGI:MGI:2157527 | Mouse | PMID:16254602 |
| Biological Process | GO:1905515 | non-motile cilium assembly | ift88 | IMP | ZFIN:ZDB-GENO-980410-147 | Zfish | PMID:15182712 |
| Biological Process | GO:0007219 | Notch signaling pathway | Ift88 | IMP | | Mouse | PMID:21703454 |
| Biological Process | GO:0007219 | Notch signaling pathway | ift88 | IMP | ZFIN:ZDB-MRPHLNO-050506-1 | Zfish | PMID:31015398 |
| Biological Process | GO:0031016 | pancreas development | Ift88 | IMP | MGI:MGI:2157527 | Mouse | PMID:11773599 |
| Biological Process | GO:0045494 | photoreceptor cell maintenance | ift88 | IMP | ZFIN:ZDB-GENO-980410-147 | Zfish | PMID:15182712 |
| Biological Process | GO:0045494 | photoreceptor cell maintenance | ift88 | IMP | ZFIN:ZDB-GENO-080730-2 | Zfish | PMID:22485131 |
| Biological Process | GO:0045724 | positive regulation of cilium assembly | IFT88 | IMP | | Human | PMID:17604723 |
| Biological Process | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway | Ift88 | IMP | | Mouse | PMID:19596798 |
| Biological Process | GO:0045880 | positive regulation of smoothened signaling pathway | ift88 | IMP | ZFIN:ZDB-GENO-110203-15 | Zfish | PMID:19700616 |
| Biological Process | GO:1903929 | primary palate development | Ift88 | IGI | MGI:MGI:1918742 | Mouse | PMID:22228099 |
| Biological Process | GO:0008104 | protein localization | Ift88 | IMP | MGI:MGI:2157527 | Mouse | PMID:22228099 |
| Biological Process | GO:0072019 | proximal convoluted tubule development | ift88 | IMP | ZFIN:ZDB-MRPHLNO-050506-2 | Zfish | PMID:19127979 |
| Biological Process | GO:0060785 | regulation of apoptosis involved in tissue homeostasis | ift88 | IMP | ZFIN:ZDB-GENO-080730-2 | Zfish | PMID:22485131 |
| Biological Process | GO:2000785 | regulation of autophagosome assembly | Ift88 | IMP | | Mouse | MGI:MGI:5527012|PMID:24089209 |
| Biological Process | GO:1902017 | regulation of cilium assembly | Ift88 | IMP | | Mouse | MGI:MGI:5527012|PMID:24089209 |
| Biological Process | GO:0045598 | regulation of fat cell differentiation | Ift88 | IMP | | Mouse | PMID:19596798 |
| Biological Process | GO:0060259 | regulation of feeding behavior | Ift88 | IMP | MGI:MGI:2182767,MGI:MGI:3710185 | Mouse | PMID:23599282 |
| Biological Process | GO:0042487 | regulation of odontogenesis of dentin-containing tooth | Ift88 | IMP | MGI:MGI:2157527 | Mouse | PMID:12701101 |
| Biological Process | GO:0070613 | regulation of protein processing | Ift88 | IMP | MGI:MGI:3027342 | Mouse | PMID:15930098 |
| Biological Process | GO:0070613 | regulation of protein processing | Ift88 | IMP | MGI:MGI:1859153 | Mouse | PMID:16254602 |
| Biological Process | GO:0034405 | response to fluid shear stress | Ift88 | IMP | MGI:MGI:2157527 | Mouse | PMID:18285569 |
| Biological Process | GO:0034021 | response to silicon dioxide | Ift88 | IEP | | Rat | PMID:32042332|RGD:155791682 |
| Biological Process | GO:0007224 | smoothened signaling pathway | Ift88 | IMP | MGI:MGI:3823090 | Mouse | PMID:19036983 |
| Biological Process | GO:0007224 | smoothened signaling pathway | Ift88 | IMP | | Mouse | PMID:19654211 |
| Biological Process | GO:0007224 | smoothened signaling pathway | Ift88 | IMP | MGI:MGI:2157527 | Mouse | PMID:22228099 |
| Biological Process | GO:0007224 | smoothened signaling pathway | Ift88 | IGI | MGI:MGI:98297 | Mouse | PMID:15930098 |
| Biological Process | GO:0007224 | smoothened signaling pathway | Ift88 | IMP | MGI:MGI:2445832,MGI:MGI:3710185 | Mouse | PMID:21429982 |
| Biological Process | GO:0007224 | smoothened signaling pathway | Ift88 | IMP | MGI:MGI:3027342 | Mouse | PMID:15930098 |
| Biological Process | GO:0007288 | sperm axoneme assembly | Ift88 | IMP | MGI:MGI:2157527 | Mouse | PMID:21337470 |
| Biological Process | GO:0007290 | spermatid nucleus elongation | Ift88 | IMP | MGI:MGI:2157527 | Mouse | PMID:21337470 |
| Biological Process | GO:0021513 | spinal cord dorsal/ventral patterning | Ift88 | IMP | MGI:MGI:1859153 | Mouse | PMID:21761479 |
| Biological Process | GO:0021537 | telencephalon development | Ift88 | IMP | MGI:MGI:3823090 | Mouse | PMID:19036983 |
 Analysis Tools
Analysis Tools