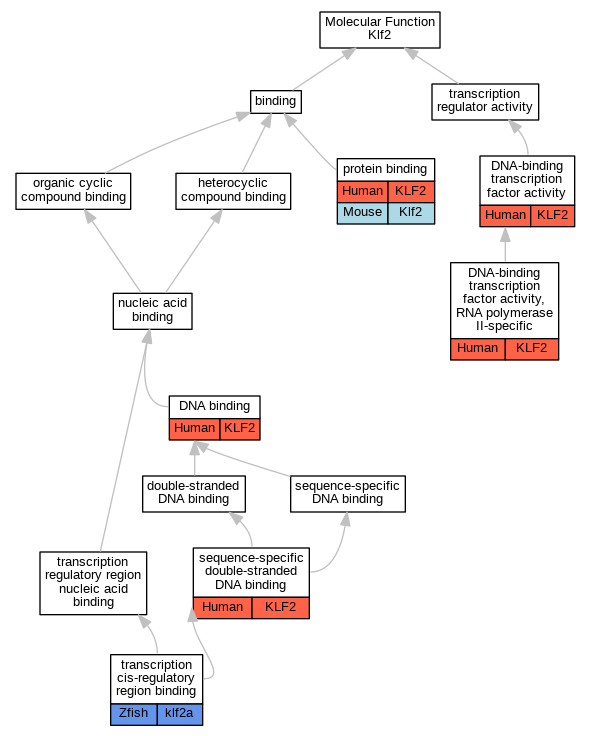
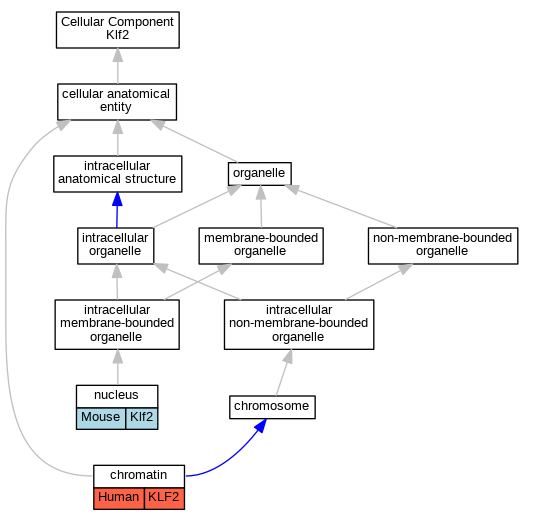
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0003677 | DNA binding | KLF2 | IMP | | Human | PMID:22327366 |
| Molecular Function | GO:0003700 | DNA-binding transcription factor activity | KLF2 | IDA | | Human | PMID:21063504 |
| Molecular Function | GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | KLF2 | IMP | | Human | PMID:22327366 |
| Molecular Function | GO:0005515 | protein binding | KLF2 | IPI | UniProtKB:Q92831 | Human | PMID:16617118 |
| Molecular Function | GO:0005515 | protein binding | KLF2 | IPI | UniProtKB:Q9HCE7 | Human | PMID:21382345 |
| Molecular Function | GO:0005515 | protein binding | KLF2 | IPI | UniProtKB:Q9HCE7-2 | Human | PMID:21382345 |
| Molecular Function | GO:0005515 | protein binding | Klf2 | IPI | UniProtKB:Q8BZZ3 | Mouse | MGI:MGI:3688077|PMID:11375995 |
| Molecular Function | GO:1990837 | sequence-specific double-stranded DNA binding | KLF2 | IDA | | Human | PMID:28473536 |
| Molecular Function | GO:0000976 | transcription cis-regulatory region binding | klf2a | IDA | | Zfish | PMID:21849483 |
| Cellular Component | GO:0000785 | chromatin | KLF2 | IMP | | Human | PMID:22327366 |
| Cellular Component | GO:0005634 | nucleus | Klf2 | IDA | | Mouse | PMID:22001906 |
| Biological Process | GO:0001525 | angiogenesis | klf2a | IGI | ZFIN:ZDB-MRPHLNO-141105-1,ZFIN:ZDB-MRPHLNO-141105-4 | Zfish | PMID:23507969 |
| Biological Process | GO:0003171 | atrioventricular valve development | klf2a | IMP | ZFIN:ZDB-MRPHLNO-101105-1 | Zfish | PMID:19924233 |
| Biological Process | GO:0003171 | atrioventricular valve development | klf2a | IMP | ZFIN:ZDB-MRPHLNO-101105-2 | Zfish | PMID:19924233 |
| Biological Process | GO:0003181 | atrioventricular valve morphogenesis | klf2a | IMP | ZFIN:ZDB-GENO-161205-6 | Zfish | PMID:27221222 |
| Biological Process | GO:0000902 | cell morphogenesis | Klf2 | IMP | MGI:MGI:3047647 | Mouse | PMID:15947087 |
| Biological Process | GO:0071409 | cellular response to cycloheximide | Klf2 | IEP | | Rat | PMID:18406357|RGD:2316543 |
| Biological Process | GO:0071498 | cellular response to fluid shear stress | KLF2 | IMP | | Human | PMID:22327366 |
| Biological Process | GO:0070301 | cellular response to hydrogen peroxide | Klf2 | IEP | | Rat | PMID:18406357|RGD:2316543 |
| Biological Process | GO:0071347 | cellular response to interleukin-1 | Klf2 | IEP | | Rat | PMID:18406357|RGD:2316543 |
| Biological Process | GO:0071499 | cellular response to laminar fluid shear stress | KLF2 | IMP | | Human | PMID:23043106 |
| Biological Process | GO:0071407 | cellular response to organic cyclic compound | Klf2 | IEP | | Rat | PMID:18406357|RGD:2316543 |
| Biological Process | GO:1901653 | cellular response to peptide | Klf2 | IEP | | Rat | PMID:18406357|RGD:2316543 |
| Biological Process | GO:0071356 | cellular response to tumor necrosis factor | Klf2 | IEP | | Rat | PMID:18406357|RGD:2316543 |
| Biological Process | GO:0097533 | cellular stress response to acid chemical | KLF2 | IMP | | Human | PMID:26299712 |
| Biological Process | GO:0061444 | endocardial cushion cell development | klf2a | IMP | ZFIN:ZDB-MRPHLNO-101105-2 | Zfish | PMID:19924233 |
| Biological Process | GO:0061444 | endocardial cushion cell development | klf2a | IMP | ZFIN:ZDB-MRPHLNO-101105-1 | Zfish | PMID:19924233 |
| Biological Process | GO:0040029 | epigenetic regulation of gene expression | Klf2 | IGI | MGI:MGI:3588194 | Mouse | PMID:22482507 |
| Biological Process | GO:0034101 | erythrocyte homeostasis | Klf2 | IMP | MGI:MGI:3047647 | Mouse | PMID:15947087 |
| Biological Process | GO:0043249 | erythrocyte maturation | Klf2 | IMP | MGI:MGI:3047647 | Mouse | PMID:15947087 |
| Biological Process | GO:0007507 | heart development | klf2a | IGI | ZFIN:ZDB-MRPHLNO-220309-1,ZFIN:ZDB-MRPHLNO-121102-1,ZFIN:ZDB-MRPHLNO-220309-2 | Zfish | PMID:25625206 |
| Biological Process | GO:0007507 | heart development | klf2b | IGI | ZFIN:ZDB-MRPHLNO-220309-1,ZFIN:ZDB-MRPHLNO-121102-1,ZFIN:ZDB-MRPHLNO-220309-2 | Zfish | PMID:25625206 |
| Biological Process | GO:0001701 | in utero embryonic development | Klf2 | IMP | MGI:MGI:3047647 | Mouse | PMID:9859212 |
| Biological Process | GO:0001701 | in utero embryonic development | Klf2 | IMP | MGI:MGI:3047647 | Mouse | PMID:15947087 |
| Biological Process | GO:0035264 | multicellular organism growth | Klf2 | IMP | MGI:MGI:3047647 | Mouse | PMID:15947087 |
| Biological Process | GO:0032715 | negative regulation of interleukin-6 production | Klf2 | IMP | | Rat | PMID:19192484|RGD:2316542 |
| Biological Process | GO:0042662 | negative regulation of mesodermal cell fate specification | klf2a | IGI | ZFIN:ZDB-MRPHLNO-100610-8,ZFIN:ZDB-MRPHLNO-140217-1 | Zfish | PMID:24211655 |
| Biological Process | GO:0042662 | negative regulation of mesodermal cell fate specification | klf2b | IGI | ZFIN:ZDB-MRPHLNO-100610-8,ZFIN:ZDB-MRPHLNO-140217-1 | Zfish | PMID:24211655 |
| Biological Process | GO:1903671 | negative regulation of sprouting angiogenesis | KLF2 | IMP | | Human | PMID:26299712 |
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | KLF2 | IDA | | Human | PMID:26299712 |
| Biological Process | GO:0006809 | nitric oxide biosynthetic process | klf2a | IMP | ZFIN:ZDB-MRPHLNO-100610-8 | Zfish | PMID:21849483 |
| Biological Process | GO:0045893 | positive regulation of DNA-templated transcription | KLF2 | IDA | | Human | PMID:21063504 |
| Biological Process | GO:0045893 | positive regulation of DNA-templated transcription | Klf2 | IDA | | Mouse | PMID:7565748 |
| Biological Process | GO:0045893 | positive regulation of DNA-templated transcription | Klf2 | IMP | | Rat | PMID:15540987|RGD:2316547 |
| Biological Process | GO:0045429 | positive regulation of nitric oxide biosynthetic process | KLF2 | IDA | | Human | PMID:21768538 |
| Biological Process | GO:0051247 | positive regulation of protein metabolic process | KLF2 | IGI | UniProtKB:Q62862 | Human | PMID:20551324 |
| Biological Process | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway | KLF2 | IDA | | Human | PMID:26416422 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | KLF2 | IGI | UniProtKB:Q62862 | Human | PMID:20551324 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | KLF2 | IMP | | Human | PMID:22327366 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | Klf2 | IMP | MGI:MGI:3047647 | Mouse | PMID:15947087 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | Klf2 | IDA | | Mouse | PMID:22001906 |
| Biological Process | GO:0036003 | positive regulation of transcription from RNA polymerase II promoter in response to stress | KLF2 | IMP | | Human | PMID:23043106 |
| Biological Process | GO:0042665 | regulation of ectodermal cell fate specification | klf2a | IGI | ZFIN:ZDB-MRPHLNO-100610-8,ZFIN:ZDB-MRPHLNO-140217-1 | Zfish | PMID:24211655 |
| Biological Process | GO:0042665 | regulation of ectodermal cell fate specification | klf2b | IGI | ZFIN:ZDB-MRPHLNO-100610-8,ZFIN:ZDB-MRPHLNO-140217-1 | Zfish | PMID:24211655 |
| Biological Process | GO:0034616 | response to laminar fluid shear stress | Klf2 | IEP | | Rat | PMID:16466697|RGD:2316544 |
| Biological Process | GO:0060509 | type I pneumocyte differentiation | Klf2 | IMP | | Mouse | PMID:22001906 |
| Biological Process | GO:0042311 | vasodilation | KLF2 | IMP | | Human | PMID:21768538 |
| Biological Process | GO:0003223 | ventricular compact myocardium morphogenesis | klf2a | IGI | ZFIN:ZDB-GENO-190606-27 | Zfish | PMID:30592462 |
| Biological Process | GO:0003223 | ventricular compact myocardium morphogenesis | klf2b | IGI | ZFIN:ZDB-GENO-190606-27 | Zfish | PMID:30592462 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
 Analysis Tools
Analysis Tools