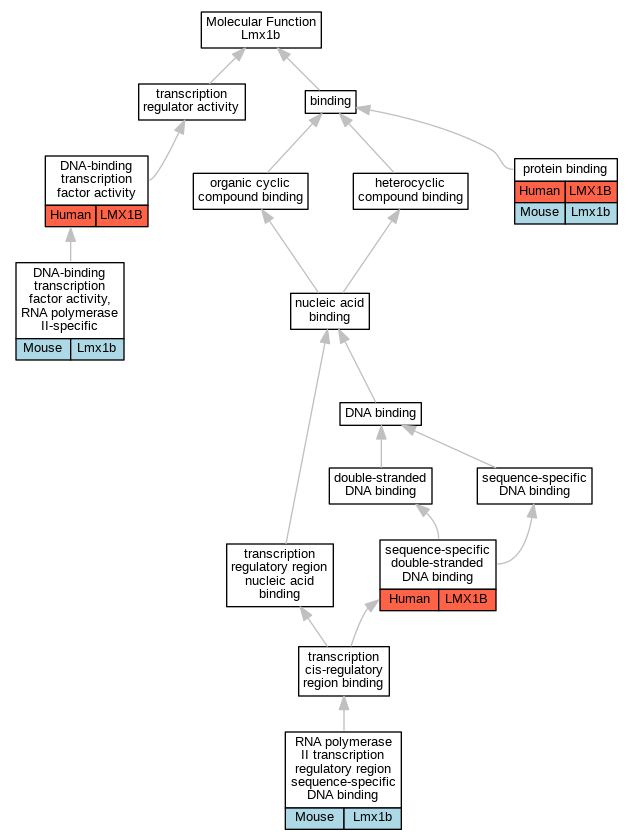
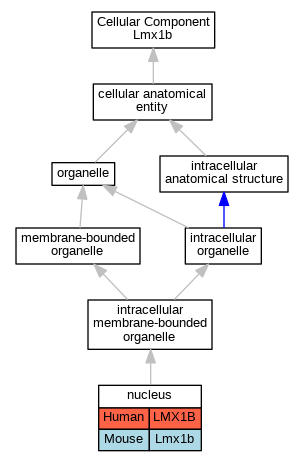
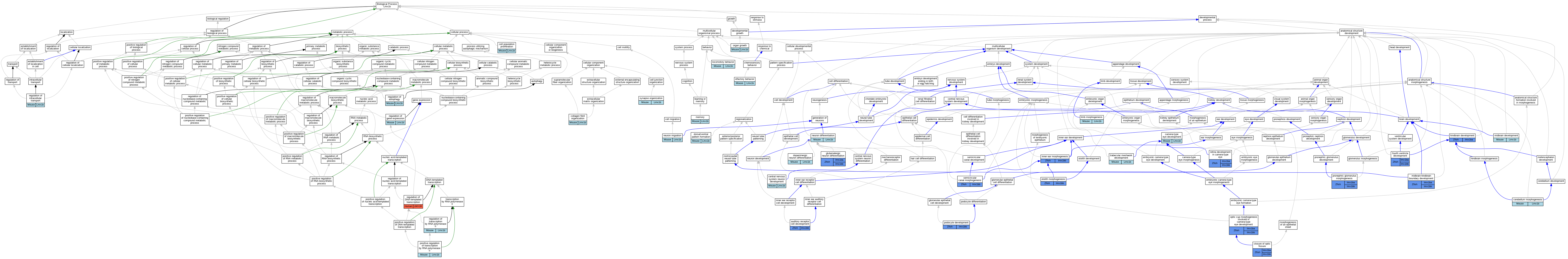
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0003700 | DNA-binding transcription factor activity | LMX1B | IDA | | Human | PMID:10767331 |
| Molecular Function | GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | Lmx1b | IDA | | Mouse | MGI:MGI:5705221|PMID:19951692 |
| Molecular Function | GO:0005515 | protein binding | LMX1B | IPI | UniProtKB:Q86U70 | Human | PMID:12792813 |
| Molecular Function | GO:0005515 | protein binding | Lmx1b | IPI | UniProtKB:Q8R326|UniProtKB:Q8VIJ6 | Mouse | MGI:MGI:5905264|PMID:23308148 |
| Molecular Function | GO:0000977 | RNA polymerase II transcription regulatory region sequence-specific DNA binding | Lmx1b | IDA | | Mouse | MGI:MGI:5705221|PMID:19951692 |
| Molecular Function | GO:1990837 | sequence-specific double-stranded DNA binding | LMX1B | IDA | | Human | PMID:28473536 |
| Cellular Component | GO:0005634 | nucleus | LMX1B | IDA | | Human | PMID:10767331 |
| Cellular Component | GO:0005634 | nucleus | LMX1B | IDA | | Human | PMID:24399192 |
| Cellular Component | GO:0005634 | nucleus | Lmx1b | IDA | | Mouse | PMID:18076286 |
| Biological Process | GO:0060117 | auditory receptor cell development | lmx1bb | IMP | ZFIN:ZDB-GENO-071114-16 | Zfish | PMID:17574823 |
| Biological Process | GO:0043010 | camera-type eye development | Lmx1b | IMP | MGI:MGI:2158463 | Mouse | PMID:10660670 |
| Biological Process | GO:0043010 | camera-type eye development | Lmx1b | IMP | MGI:MGI:2158463,MGI:MGI:2386570,MGI:MGI:3714802 | Mouse | PMID:20568247 |
| Biological Process | GO:0008283 | cell population proliferation | Lmx1b | IMP | MGI:MGI:2158463 | Mouse | PMID:17166916 |
| Biological Process | GO:0021954 | central nervous system neuron development | Lmx1b | IMP | MGI:MGI:3696968,MGI:MGI:3696982 | Mouse | PMID:17151281 |
| Biological Process | GO:0021587 | cerebellum morphogenesis | Lmx1b | IMP | MGI:MGI:2158463 | Mouse | PMID:17166916 |
| Biological Process | GO:0061386 | closure of optic fissure | lmx1bb | IGI | ZFIN:ZDB-MRPHLNO-091218-7,ZFIN:ZDB-MRPHLNO-100629-3 | Zfish | PMID:19500562 |
| Biological Process | GO:0061386 | closure of optic fissure | lmx1ba | IGI | ZFIN:ZDB-MRPHLNO-091218-7,ZFIN:ZDB-MRPHLNO-100629-3 | Zfish | PMID:19500562 |
| Biological Process | GO:0030199 | collagen fibril organization | Lmx1b | IMP | MGI:MGI:2158463 | Mouse | PMID:10660670 |
| Biological Process | GO:0071542 | dopaminergic neuron differentiation | Lmx1b | IGI | UniProtKB:Q9JKU8 | Mouse | MGI:MGI:5705221|PMID:19951692 |
| Biological Process | GO:0071542 | dopaminergic neuron differentiation | Lmx1b | IMP | MGI:MGI:2158463 | Mouse | PMID:21752929 |
| Biological Process | GO:0009953 | dorsal/ventral pattern formation | Lmx1b | IEP | | Mouse | MGI:MGI:3687803|PMID:10767331 |
| Biological Process | GO:0021592 | fourth ventricle development | lmx1bb | IGI | ZFIN:ZDB-MRPHLNO-050824-3,ZFIN:ZDB-MRPHLNO-050824-4 | Zfish | PMID:18191121 |
| Biological Process | GO:0021592 | fourth ventricle development | lmx1ba | IMP | ZFIN:ZDB-MRPHLNO-050824-4 | Zfish | PMID:18191121 |
| Biological Process | GO:1905962 | glutamatergic neuron differentiation | lmx1bb | IMP | | Zfish | PMID:27553035 |
| Biological Process | GO:1905962 | glutamatergic neuron differentiation | lmx1ba | IMP | | Zfish | PMID:27553035 |
| Biological Process | GO:0030902 | hindbrain development | lmx1ba | IMP | ZFIN:ZDB-MRPHLNO-050824-4 | Zfish | PMID:18191121 |
| Biological Process | GO:0042472 | inner ear morphogenesis | lmx1bb | IMP | ZFIN:ZDB-GENO-071114-16 | Zfish | PMID:17574823 |
| Biological Process | GO:0042472 | inner ear morphogenesis | lmx1bb | IMP | ZFIN:ZDB-GENO-071114-16 | Zfish | PMID:23052906 |
| Biological Process | GO:0042472 | inner ear morphogenesis | lmx1bb | IMP | ZFIN:ZDB-MRPHLNO-091218-7 | Zfish | PMID:23052906 |
| Biological Process | GO:0035108 | limb morphogenesis | Lmx1b | IMP | MGI:MGI:2158463 | Mouse | PMID:20589901 |
| Biological Process | GO:0007626 | locomotory behavior | Lmx1b | IGI | UniProtKB:Q9JKU8 | Mouse | MGI:MGI:5645828|PMID:25915474 |
| Biological Process | GO:0007613 | memory | Lmx1b | IGI | UniProtKB:Q9JKU8 | Mouse | MGI:MGI:5645828|PMID:25915474 |
| Biological Process | GO:0030901 | midbrain development | Lmx1b | IMP | MGI:MGI:2158463 | Mouse | PMID:21752929 |
| Biological Process | GO:0030917 | midbrain-hindbrain boundary development | lmx1bb | IGI | ZFIN:ZDB-MRPHLNO-050824-3,ZFIN:ZDB-MRPHLNO-050824-4 | Zfish | PMID:15944182 |
| Biological Process | GO:0030917 | midbrain-hindbrain boundary development | lmx1ba | IGI | ZFIN:ZDB-MRPHLNO-050824-3,ZFIN:ZDB-MRPHLNO-050824-4 | Zfish | PMID:15944182 |
| Biological Process | GO:0030182 | neuron differentiation | Lmx1b | IMP | MGI:MGI:2158463 | Mouse | PMID:12897786 |
| Biological Process | GO:0030182 | neuron differentiation | Lmx1b | IMP | MGI:MGI:2158463 | Mouse | PMID:15229182 |
| Biological Process | GO:0001764 | neuron migration | Lmx1b | IMP | MGI:MGI:2158463 | Mouse | PMID:15229182 |
| Biological Process | GO:0042048 | olfactory behavior | Lmx1b | IGI | UniProtKB:Q9JKU8 | Mouse | MGI:MGI:5645828|PMID:25915474 |
| Biological Process | GO:0002072 | optic cup morphogenesis involved in camera-type eye development | lmx1bb | IGI | ZFIN:ZDB-MRPHLNO-091218-7,ZFIN:ZDB-MRPHLNO-100629-3 | Zfish | PMID:19500562 |
| Biological Process | GO:0002072 | optic cup morphogenesis involved in camera-type eye development | lmx1ba | IGI | ZFIN:ZDB-MRPHLNO-091218-7,ZFIN:ZDB-MRPHLNO-100629-3 | Zfish | PMID:19500562 |
| Biological Process | GO:0035265 | organ growth | Lmx1b | IMP | MGI:MGI:2158463 | Mouse | PMID:17166916 |
| Biological Process | GO:0032474 | otolith morphogenesis | lmx1bb | IMP | ZFIN:ZDB-GENO-071114-16 | Zfish | PMID:17574823 |
| Biological Process | GO:0072015 | podocyte development | lmx1bb | IGI | ZFIN:ZDB-MRPHLNO-050824-3,ZFIN:ZDB-MRPHLNO-061111-2 | Zfish | PMID:24854274 |
| Biological Process | GO:0072015 | podocyte development | lmx1bb | IMP | ZFIN:ZDB-MRPHLNO-050824-3 | Zfish | PMID:24854274 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | Lmx1b | IDA | | Mouse | MGI:MGI:5705221|PMID:19951692 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | Lmx1b | IMP | | Mouse | MGI:MGI:5645828|PMID:25915474 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | Lmx1b | IMP | MGI:MGI:2158463 | Mouse | PMID:17166916 |
| Biological Process | GO:0035775 | pronephric glomerulus morphogenesis | lmx1bb | IGI | ZFIN:ZDB-MRPHLNO-050824-3,ZFIN:ZDB-MRPHLNO-050824-4,ZFIN:ZDB-MRPHLNO-140317-1 | Zfish | PMID:23990680 |
| Biological Process | GO:0035775 | pronephric glomerulus morphogenesis | lmx1bb | IGI | ZFIN:ZDB-MRPHLNO-050824-3,ZFIN:ZDB-MRPHLNO-050824-4 | Zfish | PMID:23990680 |
| Biological Process | GO:0035775 | pronephric glomerulus morphogenesis | lmx1ba | IGI | ZFIN:ZDB-MRPHLNO-050824-3,ZFIN:ZDB-MRPHLNO-050824-4,ZFIN:ZDB-MRPHLNO-140317-1 | Zfish | PMID:23990680 |
| Biological Process | GO:0035775 | pronephric glomerulus morphogenesis | lmx1ba | IGI | ZFIN:ZDB-MRPHLNO-050824-3,ZFIN:ZDB-MRPHLNO-050824-4 | Zfish | PMID:23990680 |
| Biological Process | GO:0010506 | regulation of autophagy | Lmx1b | IMP | | Mouse | MGI:MGI:5645828|PMID:25915474 |
| Biological Process | GO:0006355 | regulation of DNA-templated transcription | LMX1B | IDA | | Human | PMID:10767331 |
| Biological Process | GO:0010468 | regulation of gene expression | Lmx1b | IMP | MGI:MGI:2158463 | Mouse | PMID:20589901 |
| Biological Process | GO:0032386 | regulation of intracellular transport | Lmx1b | IMP | | Mouse | MGI:MGI:5645828|PMID:25915474 |
| Biological Process | GO:0006357 | regulation of transcription by RNA polymerase II | Lmx1b | IDA | | Mouse | PMID:21752929 |
| Biological Process | GO:0060041 | retina development in camera-type eye | lmx1bb | IGI | ZFIN:ZDB-MRPHLNO-091218-7,ZFIN:ZDB-MRPHLNO-100629-3 | Zfish | PMID:19500562 |
| Biological Process | GO:0060041 | retina development in camera-type eye | lmx1ba | IGI | ZFIN:ZDB-MRPHLNO-091218-7,ZFIN:ZDB-MRPHLNO-100629-3 | Zfish | PMID:19500562 |
| Biological Process | GO:0048752 | semicircular canal morphogenesis | lmx1bb | IMP | ZFIN:ZDB-GENO-071114-16 | Zfish | PMID:17574823 |
| Biological Process | GO:0050808 | synapse organization | Lmx1b | IGI | UniProtKB:Q9JKU8 | Mouse | MGI:MGI:5645828|PMID:25915474 |
| Biological Process | GO:0002930 | trabecular meshwork development | Lmx1b | IMP | MGI:MGI:2158463,MGI:MGI:2386570,MGI:MGI:3714802 | Mouse | PMID:20568247 |
 Analysis Tools
Analysis Tools