|
Source |
Alliance of Genome Resources |
| Category | ID | Classification term | Gene | Evidence | Inferred from | Organism | Reference |
|---|---|---|---|---|---|---|---|
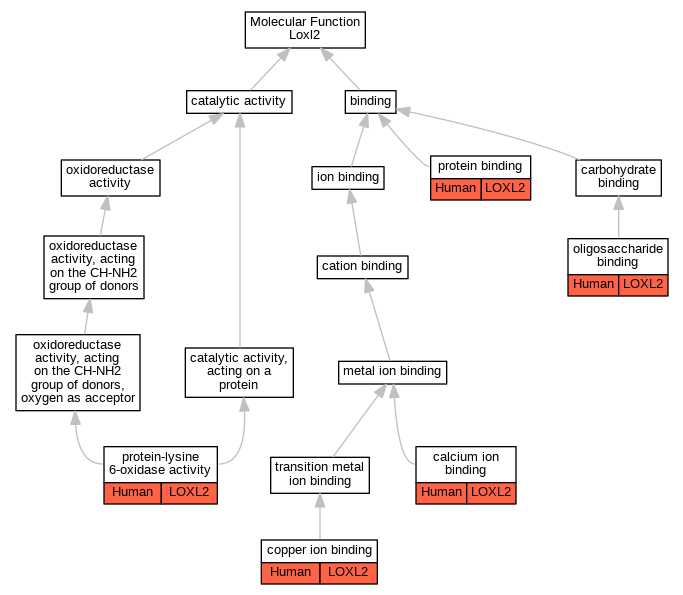
| Molecular Function | GO:0005509 | calcium ion binding | LOXL2 | IDA | Human | PMID:29581294 | |
| Molecular Function | GO:0005507 | copper ion binding | LOXL2 | IDA | Human | PMID:29581294 | |
| Molecular Function | GO:0070492 | oligosaccharide binding | LOXL2 | IDA | Human | PMID:23319596 | |
| Molecular Function | GO:0005515 | protein binding | LOXL2 | IPI | UniProtKB:O95863 | Human | PMID:16096638 |
| Molecular Function | GO:0005515 | protein binding | LOXL2 | IPI | UniProtKB:P49006 | Human | PMID:24863880 |
| Molecular Function | GO:0005515 | protein binding | LOXL2 | IPI | UniProtKB:O95967 | Human | PMID:27339457 |
| Molecular Function | GO:0005515 | protein binding | LOXL2 | IPI | UniProtKB:P15502-2 | Human | PMID:30676771 |
| Molecular Function | GO:0005515 | protein binding | LOXL2 | IPI | UniProtKB:P02751 | Human | PMID:31759052 |
| Molecular Function | GO:0004720 | protein-lysine 6-oxidase activity | LOXL2 | IDA | Human | PMID:23319596 | |
| Molecular Function | GO:0004720 | protein-lysine 6-oxidase activity | LOXL2 | IDA | Human | PMID:25959397 | |
| Molecular Function | GO:0004720 | protein-lysine 6-oxidase activity | LOXL2 | IDA | Human | PMID:27735137 | |
| Molecular Function | GO:0004720 | protein-lysine 6-oxidase activity | LOXL2 | IDA | Human | PMID:29581294 | |
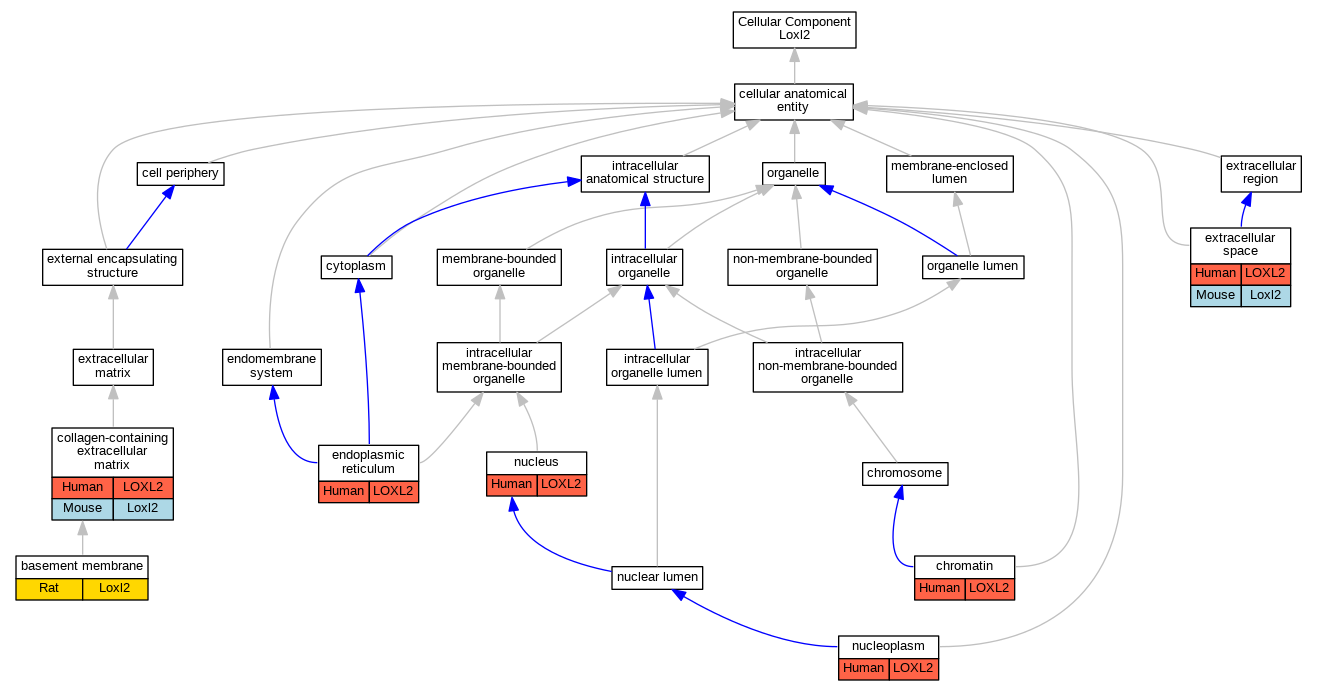
| Cellular Component | GO:0005604 | basement membrane | Loxl2 | IDA | Rat | PMID:21835952|RGD:8553848 | |
| Cellular Component | GO:0000785 | chromatin | LOXL2 | IDA | Human | PMID:27735137 | |
| Cellular Component | GO:0062023 | collagen-containing extracellular matrix | LOXL2 | HDA | Human | PMID:28327460 | |
| Cellular Component | GO:0062023 | collagen-containing extracellular matrix | LOXL2 | HDA | Human | PMID:23979707 | |
| Cellular Component | GO:0062023 | collagen-containing extracellular matrix | Loxl2 | HDA | Mouse | MGI:MGI:6201853|PMID:28071719 | |
| Cellular Component | GO:0005783 | endoplasmic reticulum | LOXL2 | IDA | Human | PMID:28332555 | |
| Cellular Component | GO:0005615 | extracellular space | LOXL2 | IDA | Human | PMID:23319596 | |
| Cellular Component | GO:0005615 | extracellular space | Loxl2 | HDA | Mouse | MGI:MGI:5640941|PMID:24006456 | |
| Cellular Component | GO:0005654 | nucleoplasm | LOXL2 | IDA | Human | GO_REF:0000052 | |
| Cellular Component | GO:0005634 | nucleus | LOXL2 | IDA | Human | PMID:22204712 | |
| Cellular Component | GO:0005634 | nucleus | LOXL2 | IDA | Human | PMID:24414204 | |
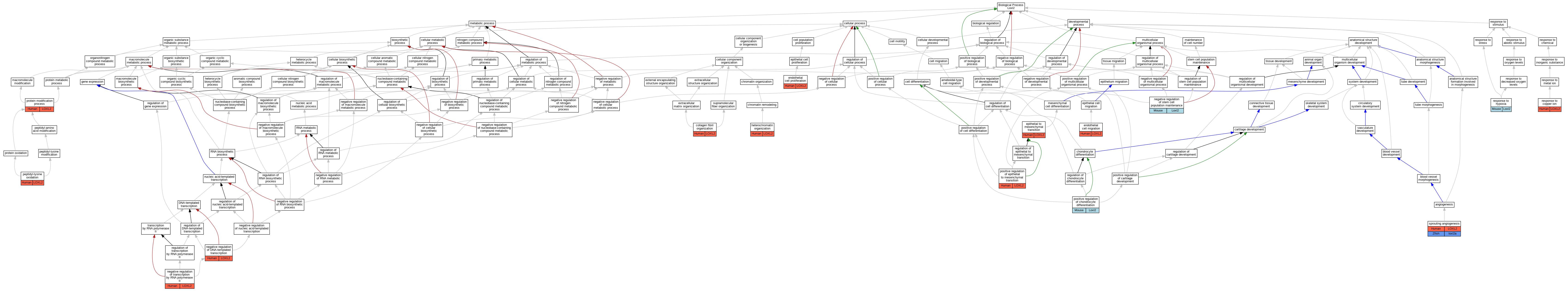
| Biological Process | GO:0030199 | collagen fibril organization | LOXL2 | IMP | Human | PMID:21835952 | |
| Biological Process | GO:0043542 | endothelial cell migration | LOXL2 | IMP | Human | PMID:21835952 | |
| Biological Process | GO:0001935 | endothelial cell proliferation | LOXL2 | IMP | Human | PMID:21835952 | |
| Biological Process | GO:0001837 | epithelial to mesenchymal transition | LOXL2 | IDA | Human | PMID:16096638 | |
| Biological Process | GO:0070828 | heterochromatin organization | LOXL2 | IMP | Human | PMID:24239292 | |
| Biological Process | GO:0045892 | negative regulation of DNA-templated transcription | LOXL2 | IDA | Human | PMID:16096638 | |
| Biological Process | GO:1902455 | negative regulation of stem cell population maintenance | Loxl2 | IDA | Mouse | MGI:MGI:6107312|PMID:25959397 | |
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | LOXL2 | IDA | Human | PMID:25959397 | |
| Biological Process | GO:0018057 | peptidyl-lysine oxidation | LOXL2 | IDA | Human | PMID:25959397 | |
| Biological Process | GO:0018057 | peptidyl-lysine oxidation | LOXL2 | IDA | Human | PMID:27735137 | |
| Biological Process | GO:0018057 | peptidyl-lysine oxidation | LOXL2 | IDA | Human | PMID:29581294 | |
| Biological Process | GO:0032332 | positive regulation of chondrocyte differentiation | Loxl2 | IMP | Mouse | MGI:MGI:5320274|PMID:21071451 | |
| Biological Process | GO:0010718 | positive regulation of epithelial to mesenchymal transition | LOXL2 | IMP | Human | PMID:24239292 | |
| Biological Process | GO:0036211 | protein modification process | LOXL2 | IDA | Human | PMID:23319596 | |
| Biological Process | GO:0046688 | response to copper ion | LOXL2 | IDA | Human | PMID:23319596 | |
| Biological Process | GO:0001666 | response to hypoxia | Loxl2 | IDA | Mouse | MGI:MGI:5320262|PMID:21835952 | |
| Biological Process | GO:0002040 | sprouting angiogenesis | LOXL2 | IMP | Human | PMID:21835952 | |
| Biological Process | GO:0002040 | sprouting angiogenesis | loxl2a | IMP | Zfish | PMID:21835952|ZFIN:ZDB-PUB-110816-15 | |
| Biological Process | GO:0002040 | sprouting angiogenesis | loxl2a | IMP | ZFIN:ZDB-MRPHLNO-121101-2 | Zfish | PMID:21835952 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 06/12/2024 MGI 6.13 |

|
|
|
||