| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
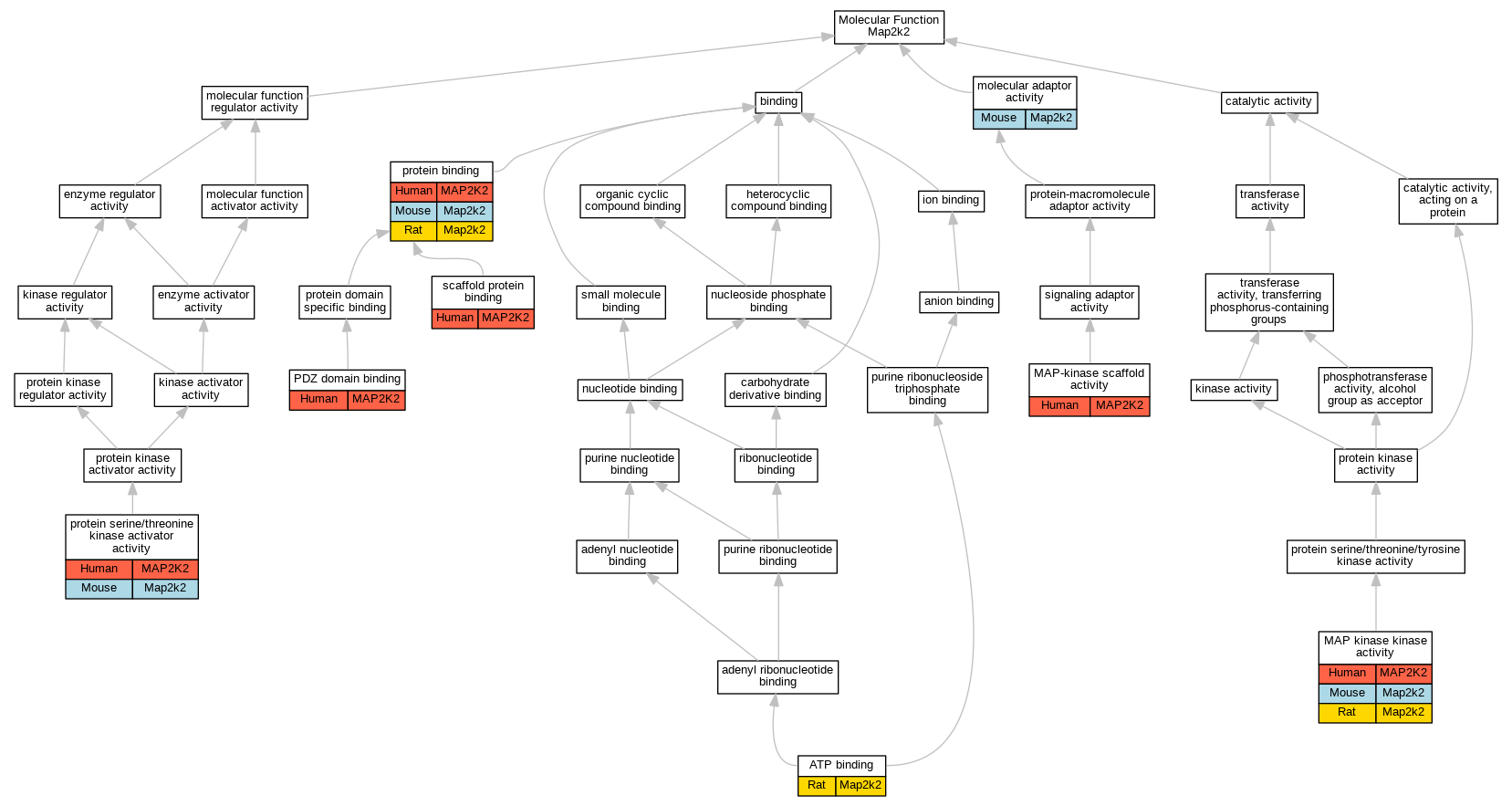
| Molecular Function | GO:0005524 | ATP binding | Map2k2 | IDA | | Rat | PMID:7611406|RGD:2298674 |
| Molecular Function | GO:0004708 | MAP kinase kinase activity | MAP2K2 | IDA | | Human | PMID:8388392 |
| Molecular Function | GO:0004708 | MAP kinase kinase activity | Map2k2 | IDA | | Mouse | PMID:8297798 |
| Molecular Function | GO:0004708 | MAP kinase kinase activity | Map2k2 | IDA | | Rat | PMID:7611406|RGD:2298674 |
| Molecular Function | GO:0004708 | MAP kinase kinase activity | Map2k2 | IDA | | Rat | PMID:8393135|RGD:61657 |
| Molecular Function | GO:0005078 | MAP-kinase scaffold activity | MAP2K2 | IMP | | Human | PMID:29433126 |
| Molecular Function | GO:0060090 | molecular adaptor activity | Map2k2 | IDA | | Mouse | MGI:MGI:5285164|PMID:20179103 |
| Molecular Function | GO:0030165 | PDZ domain binding | MAP2K2 | IDA | | Human | PMID:21615688 |
| Molecular Function | GO:0005515 | protein binding | MAP2K2 | IPI | UniProtKB:P04049 | Human | PMID:11909642 |
| Molecular Function | GO:0005515 | protein binding | MAP2K2 | IPI | UniProtKB:P10398 | Human | PMID:11909642 |
| Molecular Function | GO:0005515 | protein binding | MAP2K2 | IPI | UniProtKB:Q8IVT5 | Human | PMID:11909642 |
| Molecular Function | GO:0005515 | protein binding | MAP2K2 | IPI | UniProtKB:P15056 | Human | PMID:17979178 |
| Molecular Function | GO:0005515 | protein binding | MAP2K2 | IPI | UniProtKB:Q12959 | Human | PMID:21615688 |
| Molecular Function | GO:0005515 | protein binding | MAP2K2 | IPI | UniProtKB:P04049 | Human | PMID:21988832 |
| Molecular Function | GO:0005515 | protein binding | MAP2K2 | IPI | UniProtKB:P10398 | Human | PMID:21988832 |
| Molecular Function | GO:0005515 | protein binding | MAP2K2 | IPI | UniProtKB:P04049 | Human | PMID:24746704 |
| Molecular Function | GO:0005515 | protein binding | MAP2K2 | IPI | UniProtKB:P10398 | Human | PMID:24746704 |
| Molecular Function | GO:0005515 | protein binding | MAP2K2 | IPI | UniProtKB:P15056 | Human | PMID:24746704 |
| Molecular Function | GO:0005515 | protein binding | MAP2K2 | IPI | UniProtKB:O95273 | Human | PMID:25416956 |
| Molecular Function | GO:0005515 | protein binding | MAP2K2 | IPI | UniProtKB:P15056 | Human | PMID:25600339 |
| Molecular Function | GO:0005515 | protein binding | MAP2K2 | IPI | UniProtKB:P10398 | Human | PMID:25852190 |
| Molecular Function | GO:0005515 | protein binding | MAP2K2 | IPI | UniProtKB:Q8IVT5 | Human | PMID:27086506 |
| Molecular Function | GO:0005515 | protein binding | MAP2K2 | IPI | UniProtKB:P15056 | Human | PMID:28514442 |
| Molecular Function | GO:0005515 | protein binding | MAP2K2 | IPI | UniProtKB:P15056 | Human | PMID:29433126 |
| Molecular Function | GO:0005515 | protein binding | MAP2K2 | IPI | UniProtKB:Q8IVT5 | Human | PMID:29433126 |
| Molecular Function | GO:0005515 | protein binding | MAP2K2 | IPI | UniProtKB:P04049 | Human | PMID:31980649 |
| Molecular Function | GO:0005515 | protein binding | MAP2K2 | IPI | UniProtKB:P10398 | Human | PMID:31980649 |
| Molecular Function | GO:0005515 | protein binding | MAP2K2 | IPI | UniProtKB:P15056 | Human | PMID:31980649 |
| Molecular Function | GO:0005515 | protein binding | MAP2K2 | IPI | UniProtKB:Q8IVT5 | Human | PMID:31980649 |
| Molecular Function | GO:0005515 | protein binding | MAP2K2 | IPI | UniProtKB:P00540 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | MAP2K2 | IPI | UniProtKB:P61978-2 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | MAP2K2 | IPI | UniProtKB:Q96II5 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | MAP2K2 | IPI | UniProtKB:P04049 | Human | PMID:32707033 |
| Molecular Function | GO:0005515 | protein binding | MAP2K2 | IPI | UniProtKB:P10398 | Human | PMID:32707033 |
| Molecular Function | GO:0005515 | protein binding | MAP2K2 | IPI | UniProtKB:P15056 | Human | PMID:32707033 |
| Molecular Function | GO:0005515 | protein binding | MAP2K2 | IPI | UniProtKB:Q8IVT5 | Human | PMID:32707033 |
| Molecular Function | GO:0005515 | protein binding | MAP2K2 | IPI | UniProtKB:P05067 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | Map2k2 | IPI | UniProtKB:Q13526 | Mouse | MGI:MGI:5285164|PMID:20179103 |
| Molecular Function | GO:0005515 | protein binding | Map2k2 | IPI | UniProtKB:P85298-4|UniProtKB:Q13526 | Mouse | MGI:MGI:5285164|PMID:20179103 |
| Molecular Function | GO:0005515 | protein binding | Map2k2 | IPI | UniProtKB:Q8CGE9 | Mouse | MGI:MGI:5508605|PMID:17380122 |
| Molecular Function | GO:0005515 | protein binding | Map2k2 | IPI | RGD:1308105 | Rat | PMID:16272159|RGD:2298675 |
| Molecular Function | GO:0043539 | protein serine/threonine kinase activator activity | MAP2K2 | IDA | | Human | PMID:8388392 |
| Molecular Function | GO:0043539 | protein serine/threonine kinase activator activity | Map2k2 | IDA | | Mouse | PMID:8297798 |
| Molecular Function | GO:0097110 | scaffold protein binding | MAP2K2 | IPI | UniProtKB:Q61097 | Human | PMID:10409742 |
| Molecular Function | GO:0097110 | scaffold protein binding | MAP2K2 | IPI | UniProtKB:Q12959 | Human | PMID:21615688 |
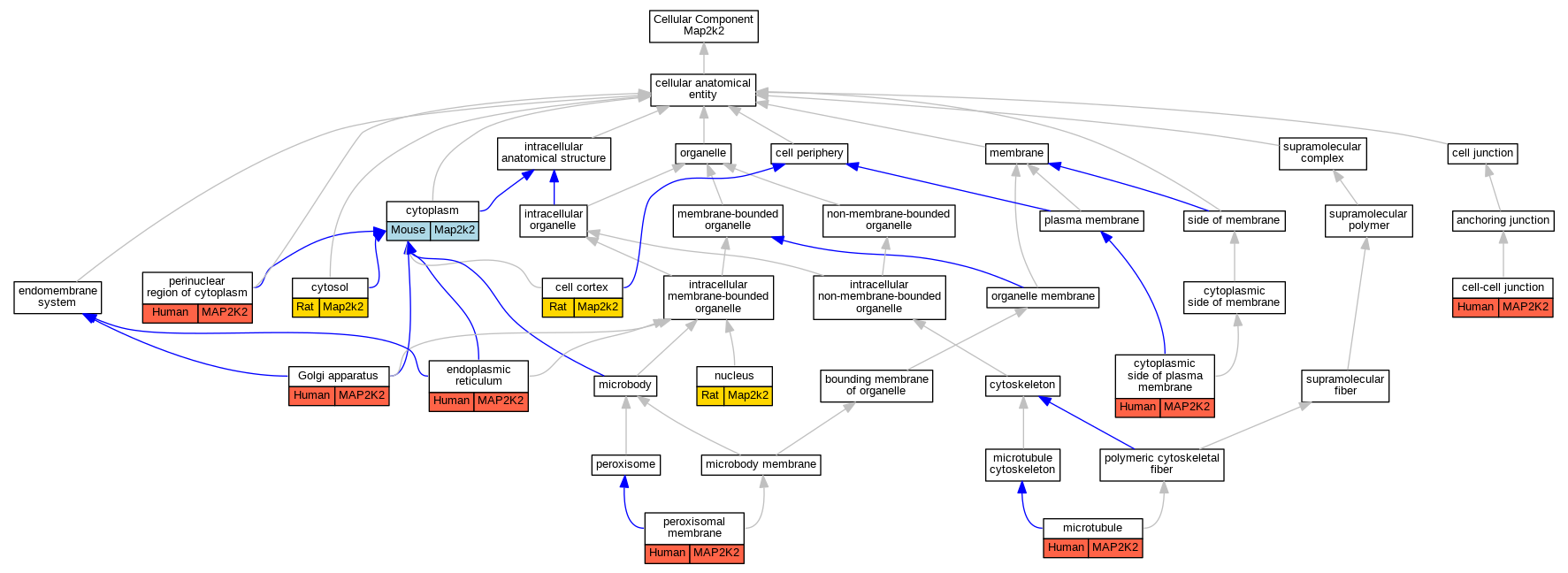
| Cellular Component | GO:0005938 | cell cortex | Map2k2 | IDA | | Rat | PMID:16272159|RGD:2298675 |
| Cellular Component | GO:0005911 | cell-cell junction | MAP2K2 | IDA | | Human | PMID:21615688 |
| Cellular Component | GO:0005737 | cytoplasm | Map2k2 | IDA | | Mouse | PMID:23920377 |
| Cellular Component | GO:0009898 | cytoplasmic side of plasma membrane | MAP2K2 | IDA | | Human | PMID:21615688 |
| Cellular Component | GO:0005829 | cytosol | Map2k2 | IDA | | Rat | PMID:16272159|RGD:2298675 |
| Cellular Component | GO:0005829 | cytosol | Map2k2 | IDA | | Rat | PMID:9169613|RGD:155791633 |
| Cellular Component | GO:0005829 | cytosol | Map2k2 | IDA | | Rat | PMID:9397988|RGD:150530477 |
| Cellular Component | GO:0005783 | endoplasmic reticulum | MAP2K2 | IDA | | Human | PMID:21615688 |
| Cellular Component | GO:0005794 | Golgi apparatus | MAP2K2 | IDA | | Human | PMID:21615688 |
| Cellular Component | GO:0005874 | microtubule | MAP2K2 | IDA | | Human | PMID:21615688 |
| Cellular Component | GO:0005634 | nucleus | Map2k2 | IDA | | Rat | PMID:9169613|RGD:155791633 |
| Cellular Component | GO:0048471 | perinuclear region of cytoplasm | MAP2K2 | IDA | | Human | PMID:21615688 |
| Cellular Component | GO:0005778 | peroxisomal membrane | MAP2K2 | HDA | | Human | PMID:21525035 |
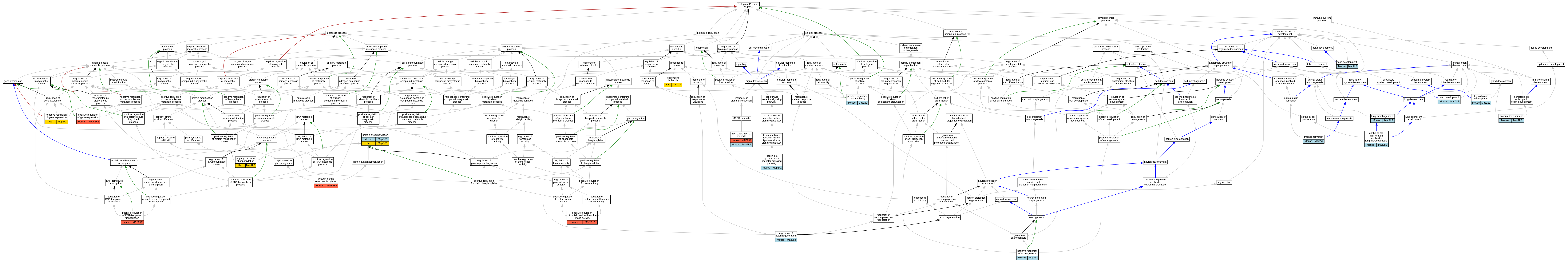
| Biological Process | GO:0060502 | epithelial cell proliferation involved in lung morphogenesis | Map2k2 | IGI | MGI:MGI:1346866 | Mouse | PMID:25100655 |
| Biological Process | GO:0070371 | ERK1 and ERK2 cascade | MAP2K2 | IMP | | Human | PMID:24375836 |
| Biological Process | GO:0070371 | ERK1 and ERK2 cascade | Map2k2 | IGI | MGI:MGI:1346866 | Mouse | PMID:25100655 |
| Biological Process | GO:0060324 | face development | Map2k2 | IGI | MGI:MGI:1346866 | Mouse | PMID:18952847 |
| Biological Process | GO:0007507 | heart development | Map2k2 | IGI | MGI:MGI:1346866 | Mouse | PMID:18952847 |
| Biological Process | GO:0048009 | insulin-like growth factor receptor signaling pathway | Map2k2 | IMP | | Mouse | PMID:21699731 |
| Biological Process | GO:0060425 | lung morphogenesis | Map2k2 | IGI | MGI:MGI:1346866 | Mouse | PMID:25100655 |
| Biological Process | GO:0010629 | negative regulation of gene expression | Map2k2 | IGI | UniProtKB:Q01986 | Rat | PMID:28947594|RGD:13524616 |
| Biological Process | GO:0036289 | peptidyl-serine autophosphorylation | MAP2K2 | IDA | | Human | PMID:8388392 |
| Biological Process | GO:0018108 | peptidyl-tyrosine phosphorylation | Map2k2 | IDA | | Rat | PMID:9169613|RGD:155791633 |
| Biological Process | GO:0050772 | positive regulation of axonogenesis | Map2k2 | IGI | MGI:MGI:1346866 | Mouse | PMID:24733831 |
| Biological Process | GO:0050772 | positive regulation of axonogenesis | Map2k2 | IGI | MGI:MGI:88190 | Mouse | PMID:24733831 |
| Biological Process | GO:2000147 | positive regulation of cell motility | Map2k2 | IDA | | Mouse | MGI:MGI:5285164|PMID:20179103 |
| Biological Process | GO:0045893 | positive regulation of DNA-templated transcription | MAP2K2 | IMP | | Human | PMID:24375836 |
| Biological Process | GO:0010628 | positive regulation of gene expression | MAP2K2 | IMP | | Human | PMID:24375836 |
| Biological Process | GO:0071902 | positive regulation of protein serine/threonine kinase activity | MAP2K2 | IDA | | Human | PMID:8388392 |
| Biological Process | GO:0006468 | protein phosphorylation | Map2k2 | IGI | MGI:MGI:1346866 | Mouse | PMID:19047410 |
| Biological Process | GO:0006468 | protein phosphorylation | Map2k2 | IDA | | Rat | PMID:7611406|RGD:2298674 |
| Biological Process | GO:0006468 | protein phosphorylation | Map2k2 | IDA | | Rat | PMID:8393135|RGD:61657 |
| Biological Process | GO:0048679 | regulation of axon regeneration | Map2k2 | IGI | MGI:MGI:1346866 | Mouse | PMID:24733831 |
| Biological Process | GO:0048679 | regulation of axon regeneration | Map2k2 | IGI | MGI:MGI:88190 | Mouse | PMID:24733831 |
| Biological Process | GO:0002931 | response to ischemia | Map2k2 | IDA | | Rat | PMID:9169613|RGD:155791633 |
| Biological Process | GO:0048538 | thymus development | Map2k2 | IGI | MGI:MGI:1346866 | Mouse | PMID:18952847 |
| Biological Process | GO:0030878 | thyroid gland development | Map2k2 | IGI | MGI:MGI:1346866 | Mouse | PMID:18952847 |
| Biological Process | GO:0060440 | trachea formation | Map2k2 | IGI | MGI:MGI:1346866 | Mouse | PMID:25100655 |
 Analysis Tools
Analysis Tools