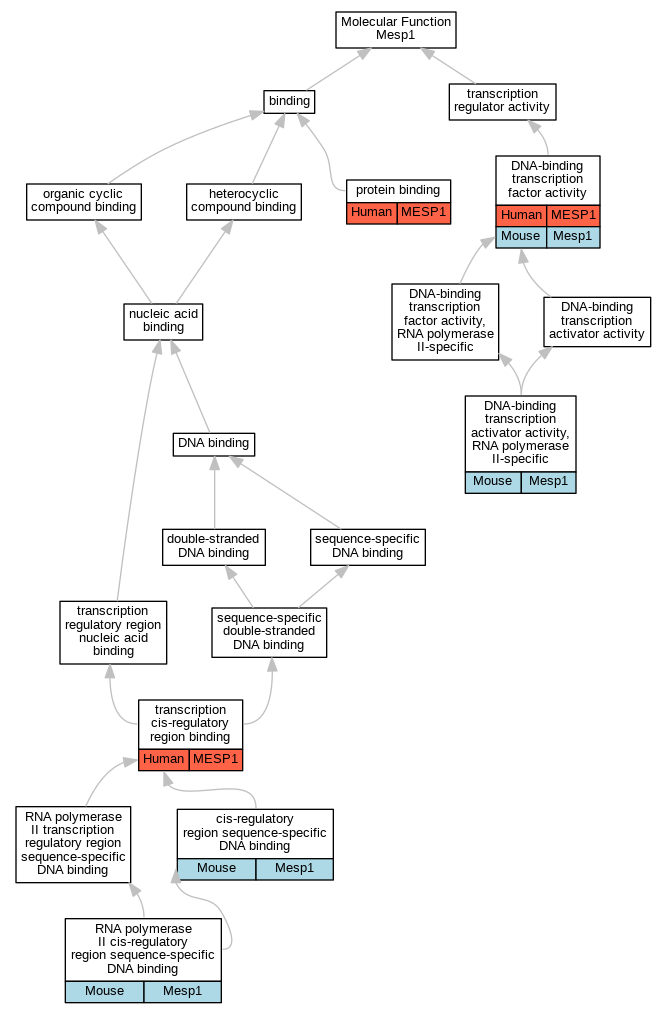
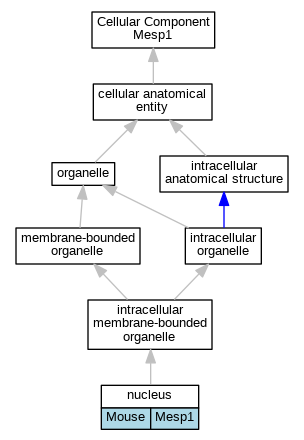
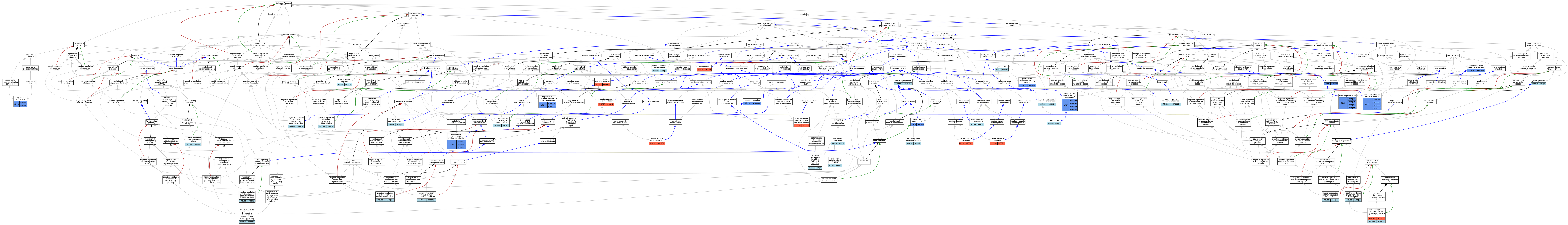
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0000987 | cis-regulatory region sequence-specific DNA binding | Mesp1 | IDA | | Mouse | MGI:MGI:4441249|PMID:18593560 |
| Molecular Function | GO:0001228 | DNA-binding transcription activator activity, RNA polymerase II-specific | Mesp1 | IDA | | Mouse | MGI:MGI:3835736|PMID:18297060 |
| Molecular Function | GO:0001228 | DNA-binding transcription activator activity, RNA polymerase II-specific | Mesp1 | IMP | | Mouse | MGI:MGI:3835736|PMID:18297060 |
| Molecular Function | GO:0003700 | DNA-binding transcription factor activity | MESP1 | IDA | | Human | PMID:18297060 |
| Molecular Function | GO:0003700 | DNA-binding transcription factor activity | Mesp1 | IMP | | Mouse | MGI:MGI:3835736|PMID:18297060 |
| Molecular Function | GO:0005515 | protein binding | MESP1 | IPI | UniProtKB:Q7Z699 | Human | PMID:32814053 |
| Molecular Function | GO:0000978 | RNA polymerase II cis-regulatory region sequence-specific DNA binding | Mesp1 | IDA | | Mouse | MGI:MGI:3835736|PMID:18297060 |
| Molecular Function | GO:0000976 | transcription cis-regulatory region binding | MESP1 | IDA | | Human | PMID:18297060 |
| Cellular Component | GO:0005634 | nucleus | Mesp1 | IDA | | Mouse | MGI:MGI:4441249|PMID:18593560 |
| Biological Process | GO:0009952 | anterior/posterior pattern specification | mespba | IMP | | Zfish | PMID:10725245 |
| Biological Process | GO:0097101 | blood vessel endothelial cell fate specification | mespaa | IMP | | Zfish | PMID:27554166 |
| Biological Process | GO:0097101 | blood vessel endothelial cell fate specification | mespba | IMP | | Zfish | PMID:27554166 |
| Biological Process | GO:0097101 | blood vessel endothelial cell fate specification | mespbb | IMP | | Zfish | PMID:27554166 |
| Biological Process | GO:0003210 | cardiac atrium formation | MESP1 | IMP | | Human | PMID:18297060 |
| Biological Process | GO:0060913 | cardiac cell fate determination | Mesp1 | IDA | | Mouse | MGI:MGI:4441249|PMID:18593560 |
| Biological Process | GO:0055007 | cardiac muscle cell differentiation | MESP1 | IMP | | Human | PMID:18297060 |
| Biological Process | GO:0060947 | cardiac vascular smooth muscle cell differentiation | MESP1 | IMP | | Human | PMID:18297060 |
| Biological Process | GO:0003211 | cardiac ventricle formation | MESP1 | IMP | | Human | PMID:18297060 |
| Biological Process | GO:0003259 | cardioblast anterior-lateral migration | Mesp1 | IEP | | Mouse | MGI:MGI:3835736|PMID:18297060 |
| Biological Process | GO:0003260 | cardioblast migration | Mesp1 | IMP | MGI:MGI:2182080 | Mouse | PMID:10393122 |
| Biological Process | GO:0060975 | cardioblast migration to the midline involved in heart field formation | Mesp1 | IMP | | Mouse | MGI:MGI:1339460|PMID:10393122 |
| Biological Process | GO:0061371 | determination of heart left/right asymmetry | mespaa | IGI | ZFIN:ZDB-GENO-161206-2,ZFIN:ZDB-MRPHLNO-161122-1 | Zfish | PMID:27554166 |
| Biological Process | GO:0061371 | determination of heart left/right asymmetry | mespaa | IGI | ZFIN:ZDB-GENO-161206-2,ZFIN:ZDB-MRPHLNO-161122-2 | Zfish | PMID:27554166 |
| Biological Process | GO:0061371 | determination of heart left/right asymmetry | mespaa | IMP | ZFIN:ZDB-GENO-161206-2 | Zfish | PMID:27554166 |
| Biological Process | GO:0061371 | determination of heart left/right asymmetry | mespaa | IGI | ZFIN:ZDB-GENO-161206-2,ZFIN:ZDB-MRPHLNO-060113-7 | Zfish | PMID:27554166 |
| Biological Process | GO:0061371 | determination of heart left/right asymmetry | mespba | IGI | ZFIN:ZDB-GENO-161206-2,ZFIN:ZDB-MRPHLNO-060113-7 | Zfish | PMID:27554166 |
| Biological Process | GO:0061371 | determination of heart left/right asymmetry | mespab | IGI | ZFIN:ZDB-GENO-161206-2,ZFIN:ZDB-MRPHLNO-161122-1 | Zfish | PMID:27554166 |
| Biological Process | GO:0061371 | determination of heart left/right asymmetry | mespbb | IGI | ZFIN:ZDB-GENO-161206-2,ZFIN:ZDB-MRPHLNO-161122-2 | Zfish | PMID:27554166 |
| Biological Process | GO:0003143 | embryonic heart tube morphogenesis | Mesp1 | IMP | | Mouse | MGI:MGI:1339460|PMID:10393122 |
| Biological Process | GO:0045446 | endothelial cell differentiation | MESP1 | IMP | | Human | PMID:18297060 |
| Biological Process | GO:0007369 | gastrulation | Mesp1 | IEP | | Mouse | MGI:MGI:83220|PMID:8787751 |
| Biological Process | GO:0001702 | gastrulation with mouth forming second | mespaa | IMP | | Zfish | PMID:10725245 |
| Biological Process | GO:0010467 | gene expression | Mesp1 | IMP | | Mouse | PMID:23352431 |
| Biological Process | GO:0003241 | growth involved in heart morphogenesis | Mesp1 | IMP | | Mouse | MGI:MGI:1339460|PMID:10393122 |
| Biological Process | GO:0003128 | heart field specification | mespaa | IMP | | Zfish | PMID:27554166 |
| Biological Process | GO:0001947 | heart looping | Mesp1 | IMP | | Mouse | MGI:MGI:1339460|PMID:10393122 |
| Biological Process | GO:0003007 | heart morphogenesis | Mesp1 | IMP | MGI:MGI:2182080 | Mouse | PMID:10393122 |
| Biological Process | GO:0048368 | lateral mesoderm development | Mesp1 | IEP | | Mouse | MGI:MGI:1339460|PMID:10393122 |
| Biological Process | GO:0001707 | mesoderm formation | mespaa | IMP | | Zfish | PMID:10725245 |
| Biological Process | GO:0008078 | mesodermal cell migration | Mesp1 | IGI | MGI:MGI:1096325 | Mouse | PMID:10393122 |
| Biological Process | GO:0045892 | negative regulation of DNA-templated transcription | Mesp1 | IDA | | Mouse | MGI:MGI:4441249|PMID:18593560 |
| Biological Process | GO:0042664 | negative regulation of endodermal cell fate specification | Mesp1 | IDA | | Mouse | MGI:MGI:4441249|PMID:18593560 |
| Biological Process | GO:0042662 | negative regulation of mesodermal cell fate specification | Mesp1 | IDA | | Mouse | MGI:MGI:4441249|PMID:18593560 |
| Biological Process | GO:0022008 | neurogenesis | MESP1 | IMP | | Human | PMID:18297060 |
| Biological Process | GO:0045893 | positive regulation of DNA-templated transcription | Mesp1 | IDA | | Mouse | MGI:MGI:4441249|PMID:18593560 |
| Biological Process | GO:0090082 | positive regulation of heart induction by negative regulation of canonical Wnt signaling pathway | Mesp1 | IDA | | Mouse | MGI:MGI:3835736|PMID:18297060 |
| Biological Process | GO:0070368 | positive regulation of hepatocyte differentiation | Mesp1 | IDA | | Mouse | MGI:MGI:4441249|PMID:18593560 |
| Biological Process | GO:0045747 | positive regulation of Notch signaling pathway | Mesp1 | IDA | | Mouse | MGI:MGI:4441249|PMID:18593560 |
| Biological Process | GO:0035481 | positive regulation of Notch signaling pathway involved in heart induction | Mesp1 | IDA | | Mouse | MGI:MGI:4441249|PMID:18593560 |
| Biological Process | GO:0051155 | positive regulation of striated muscle cell differentiation | Mesp1 | IDA | | Mouse | MGI:MGI:4441249|PMID:18593560 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | MESP1 | IDA | | Human | PMID:18297060 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | Mesp1 | IMP | | Mouse | MGI:MGI:3835736|PMID:18297060 |
| Biological Process | GO:0048641 | regulation of skeletal muscle tissue development | mespba | IGI | ZFIN:ZDB-GENO-160517-1 | Zfish | PMID:25725067 |
| Biological Process | GO:0048641 | regulation of skeletal muscle tissue development | mespba | IGI | ZFIN:ZDB-MRPHLNO-060113-7,ZFIN:ZDB-MRPHLNO-150603-3 | Zfish | PMID:25725067 |
| Biological Process | GO:0048641 | regulation of skeletal muscle tissue development | mespba | IGI | ZFIN:ZDB-GENO-160517-2 | Zfish | PMID:25725067 |
| Biological Process | GO:0048641 | regulation of skeletal muscle tissue development | mespbb | IGI | ZFIN:ZDB-MRPHLNO-060113-7,ZFIN:ZDB-MRPHLNO-150603-3 | Zfish | PMID:25725067 |
| Biological Process | GO:0032526 | response to retinoic acid | mespaa | IDA | | Zfish | PMID:18261720 |
| Biological Process | GO:0032526 | response to retinoic acid | mespba | IDA | | Zfish | PMID:18261720 |
| Biological Process | GO:0003139 | secondary heart field specification | Mesp1 | IDA | | Mouse | MGI:MGI:4441249|PMID:18593560 |
| Biological Process | GO:0023019 | signal transduction involved in regulation of gene expression | Mesp1 | IDA | | Mouse | PMID:18462699 |
| Biological Process | GO:0060921 | sinoatrial node cell differentiation | MESP1 | IMP | | Human | PMID:18297060 |
| Biological Process | GO:0003236 | sinus venosus morphogenesis | Mesp1 | IMP | | Mouse | MGI:MGI:1339460|PMID:10393122 |
| Biological Process | GO:0032525 | somite rostral/caudal axis specification | mespaa | IGI | ZFIN:ZDB-GENO-161208-3 | Zfish | PMID:27385009 |
| Biological Process | GO:0032525 | somite rostral/caudal axis specification | mespba | IGI | ZFIN:ZDB-GENO-161208-3 | Zfish | PMID:27385009 |
| Biological Process | GO:0032525 | somite rostral/caudal axis specification | mespab | IGI | ZFIN:ZDB-GENO-161208-3 | Zfish | PMID:27385009 |
| Biological Process | GO:0032525 | somite rostral/caudal axis specification | mespbb | IGI | ZFIN:ZDB-GENO-161208-3 | Zfish | PMID:27385009 |
| Biological Process | GO:0001757 | somite specification | mespaa | IGI | ZFIN:ZDB-GENO-161208-3 | Zfish | PMID:27385009 |
| Biological Process | GO:0001757 | somite specification | mespba | IGI | ZFIN:ZDB-GENO-161208-3 | Zfish | PMID:27385009 |
| Biological Process | GO:0001757 | somite specification | mespab | IGI | ZFIN:ZDB-GENO-161208-3 | Zfish | PMID:27385009 |
| Biological Process | GO:0001757 | somite specification | mespbb | IGI | ZFIN:ZDB-GENO-161208-3 | Zfish | PMID:27385009 |
| Biological Process | GO:0001756 | somitogenesis | mespba | IMP | | Zfish | PMID:10725245 |
| Biological Process | GO:0001756 | somitogenesis | mespba | IMP | ZFIN:ZDB-MRPHLNO-060113-7 | Zfish | PMID:16326386 |
 Analysis Tools
Analysis Tools