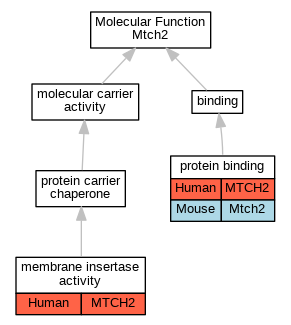
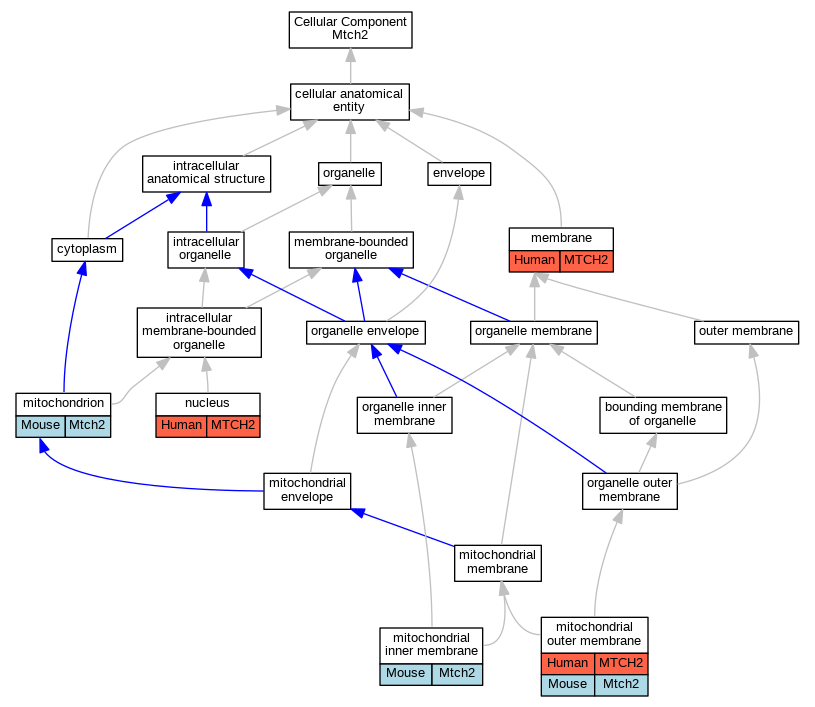
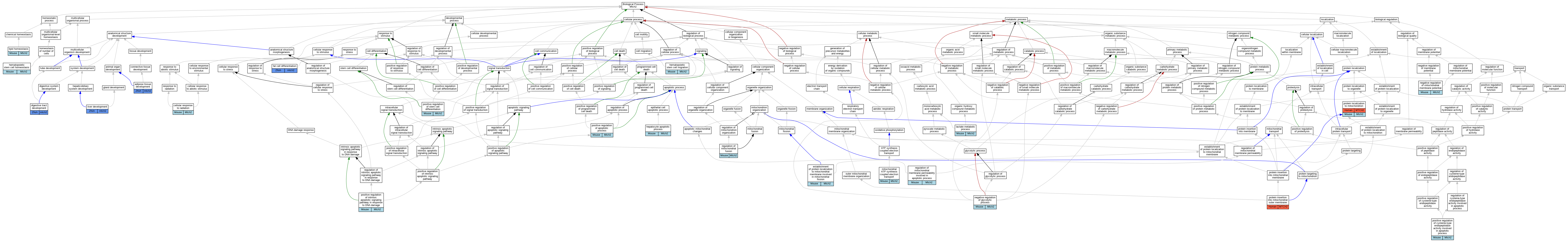
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0032977 | membrane insertase activity | MTCH2 | IDA | | Human | PMID:36264797 |
| Molecular Function | GO:0005515 | protein binding | MTCH2 | IPI | UniProtKB:P02654 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | MTCH2 | IPI | UniProtKB:P56378-2 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | Mtch2 | IPI | PR:P70444 | Mouse | PMID:20436477 |
| Cellular Component | GO:0016020 | membrane | MTCH2 | HDA | | Human | PMID:19946888 |
| Cellular Component | GO:0005743 | mitochondrial inner membrane | Mtch2 | HDA | | Mouse | PMID:12865426 |
| Cellular Component | GO:0005741 | mitochondrial outer membrane | MTCH2 | IDA | | Human | PMID:36264797 |
| Cellular Component | GO:0005741 | mitochondrial outer membrane | Mtch2 | IDA | | Mouse | PMID:20436477 |
| Cellular Component | GO:0005739 | mitochondrion | Mtch2 | HDA | | Mouse | PMID:18614015 |
| Cellular Component | GO:0005634 | nucleus | MTCH2 | HDA | | Human | PMID:21630459 |
| Biological Process | GO:0060612 | adipose tissue development | mtch2 | IMP | | Zfish | PMID:27468124|ZFIN:ZDB-PUB-160729-2 |
| Biological Process | GO:0071478 | cellular response to radiation | Mtch2 | IMP | MGI:MGI:2449949,MGI:MGI:5566664 | Mouse | PMID:26219591 |
| Biological Process | GO:0048565 | digestive tract development | mtch2 | IMP | ZFIN:ZDB-MRPHLNO-170109-4 | Zfish | PMID:27468124 |
| Biological Process | GO:0048565 | digestive tract development | mtch2 | IMP | ZFIN:ZDB-MRPHLNO-170109-3 | Zfish | PMID:27468124 |
| Biological Process | GO:0090152 | establishment of protein localization to mitochondrial membrane involved in mitochondrial fission | Mtch2 | IMP | MGI:MGI:2449949,MGI:MGI:5566664 | Mouse | PMID:26219591 |
| Biological Process | GO:0045444 | fat cell differentiation | mtch2 | IMP | ZFIN:ZDB-MRPHLNO-170109-3 | Zfish | PMID:27468124 |
| Biological Process | GO:0045444 | fat cell differentiation | mtch2 | IMP | ZFIN:ZDB-MRPHLNO-170109-4 | Zfish | PMID:27468124 |
| Biological Process | GO:0061484 | hematopoietic stem cell homeostasis | Mtch2 | IMP | MGI:MGI:2449949,MGI:MGI:5566664 | Mouse | PMID:26219591 |
| Biological Process | GO:0035701 | hematopoietic stem cell migration | Mtch2 | IMP | MGI:MGI:2449949,MGI:MGI:5566664 | Mouse | PMID:26219591 |
| Biological Process | GO:0097284 | hepatocyte apoptotic process | Mtch2 | IGI | MGI:MGI:95484 | Mouse | PMID:20436477 |
| Biological Process | GO:0097284 | hepatocyte apoptotic process | Mtch2 | IGI | MGI:MGI:108093 | Mouse | PMID:20436477 |
| Biological Process | GO:0006089 | lactate metabolic process | Mtch2 | IMP | MGI:MGI:2449949,MGI:MGI:5566664 | Mouse | PMID:26219591 |
| Biological Process | GO:0055088 | lipid homeostasis | Mtch2 | IMP | | Mouse | MGI:MGI:6814710|PMID:28127879 |
| Biological Process | GO:0001889 | liver development | mtch2 | IMP | ZFIN:ZDB-MRPHLNO-170109-3 | Zfish | PMID:27468124 |
| Biological Process | GO:0001889 | liver development | mtch2 | IMP | ZFIN:ZDB-MRPHLNO-170109-4 | Zfish | PMID:27468124 |
| Biological Process | GO:0042775 | mitochondrial ATP synthesis coupled electron transport | Mtch2 | IMP | MGI:MGI:2449949,MGI:MGI:5566664 | Mouse | PMID:26219591 |
| Biological Process | GO:0045820 | negative regulation of glycolytic process | Mtch2 | IMP | MGI:MGI:2449949,MGI:MGI:5566664 | Mouse | PMID:26219591 |
| Biological Process | GO:0010917 | negative regulation of mitochondrial membrane potential | Mtch2 | IMP | MGI:MGI:2449949,MGI:MGI:5566664 | Mouse | PMID:26219591 |
| Biological Process | GO:0043065 | positive regulation of apoptotic process | Mtch2 | IMP | | Mouse | MGI:MGI:3581108|PMID:15899861 |
| Biological Process | GO:0043280 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic process | Mtch2 | IMP | MGI:MGI:2449949,MGI:MGI:5566664 | Mouse | PMID:26219591 |
| Biological Process | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage | Mtch2 | IMP | MGI:MGI:2449949,MGI:MGI:5566664 | Mouse | PMID:26219591 |
| Biological Process | GO:2000738 | positive regulation of stem cell differentiation | Mtch2 | IMP | | Mouse | MGI:MGI:6267820|PMID:30510213 |
| Biological Process | GO:0045040 | protein insertion into mitochondrial outer membrane | MTCH2 | IDA | | Human | PMID:36264797 |
| Biological Process | GO:0070585 | protein localization to mitochondrion | MTCH2 | IMP | | Human | PMID:23744079 |
| Biological Process | GO:0070585 | protein localization to mitochondrion | Mtch2 | IMP | MGI:MGI:5566665 | Mouse | PMID:20436477 |
| Biological Process | GO:0070585 | protein localization to mitochondrion | Mtch2 | IMP | MGI:MGI:2449949,MGI:MGI:5566664 | Mouse | PMID:26219591 |
| Biological Process | GO:0070585 | protein localization to mitochondrion | Mtch2 | IMP | | Mouse | PMID:23744079 |
| Biological Process | GO:0010635 | regulation of mitochondrial fusion | Mtch2 | IMP | | Mouse | MGI:MGI:6267820|PMID:30510213 |
| Biological Process | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process | Mtch2 | IGI | MGI:MGI:108093 | Mouse | PMID:23744079 |
| Biological Process | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process | Mtch2 | IGI | MGI:MGI:99702 | Mouse | PMID:23744079 |
| Biological Process | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process | Mtch2 | IGI | MGI:MGI:108093 | Mouse | PMID:20436477 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
 Analysis Tools
Analysis Tools